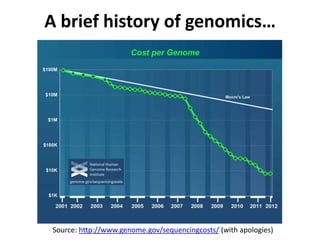
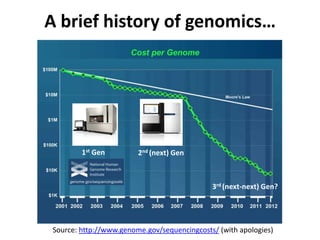
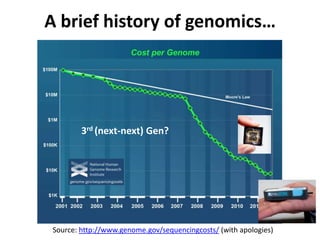
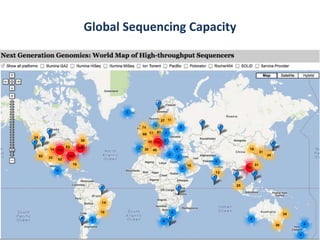
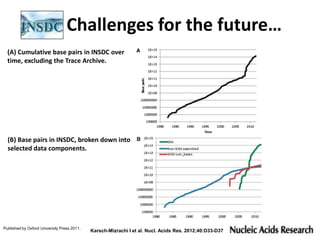
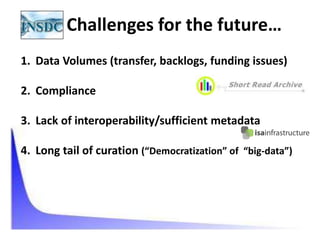
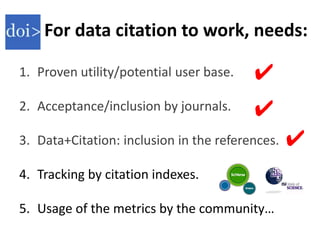
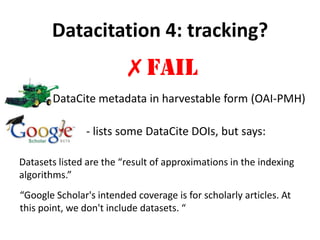
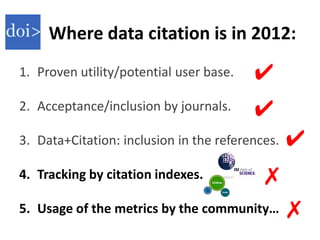
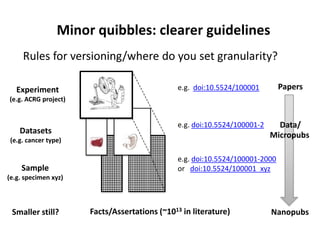
This document discusses issues around data sharing in genomics research. It provides background on the history of genomics projects like the Human Genome Project. It then discusses BGI's role in large-scale sequencing efforts and their goal of making sequencing data highly accessible. It also discusses challenges around sharing large volumes of genomic data and ensuring proper attribution and credit for data sharing. Issues around data citation are examined, including the need for data citations to be tracked by citation indexes and for metrics around data citations to be utilized by the research community.