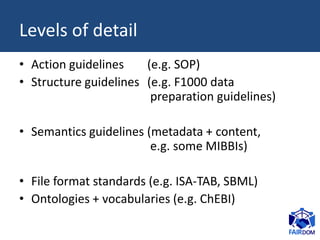
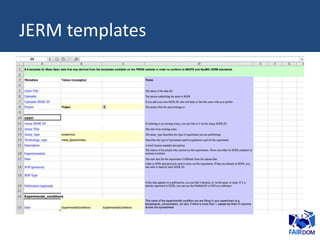
The document discusses the challenges of data sharing in the context of scientific experiments, emphasizing the importance of preparing data for dissemination while avoiding excessive effort. It proposes the 80-20 rule for maximizing benefits with minimal effort and outlines guidelines for data standardization, including metadata requirements and proper identifier management. Additionally, it suggests adopting consistent data structures and clear documentation to enhance usability across collaborative projects and fields.