This document discusses amylases, which are enzymes that degrade starch. It describes the discovery of amylases in 1833 and outlines their mechanism of breaking glycosidic bonds in starch. The document also categorizes different types of amylases based on their substrate specificity and reaction products. Finally, it reviews the industrial applications of amylases, particularly in starch processing, baking, detergents, textiles, biofuel production and paper making.

![INTRODUCTION
• Amylases are starch degrading enzymes.
• It was first isolated by French chemists Anselme and Jean François
from germinating barley and was named as "diastase" in 1833[1].
• These enzymes act by hydrolyzing glycosidic bonds-α-1,4 glycosidic
bonds and α-1,6 glycosidic bonds between adjacent glucose units,
yielding progressively smaller polymers composed of glucose units
(characteristic of the particular enzyme involved) [2] .
• They belong to the glycoside hydrolase group of enzymes under
which 13 enzymes are included [3] .
[1]- Sivaramakrishnan et al., 2006 [2]- Aiyer et al.,2005 [3]- ] Windish et al.,2005](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-2-320.jpg)
![MAJOR SUBSTRATE FOR AMYLASE -STARCH
Starch is a polymer of glucose
linked to another one through the
glycoside bond. Two types of
glucose polymers are present in
starch : amylose and amylopectin.
.
• Amylose is a linear polymer
consisting of up to 6000 glucose
units with α-1,4 glycosidic
bonds.
• Amylopectin consists of short α-
1,4 linked to linear chains of
10–60 glucose units and α-1,6
linked to side chains with 15–
45 glucose units.
• Amylase is able to cleave these
glycosidic bonds present in the
inner part of the amylose or
amylopectin chain[4].
[4] Muralikrishna G., Nirmala M. Cereal α-amylases—an
overview.Carbohydrate Polymers. 2005;60:163–173.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-4-320.jpg)
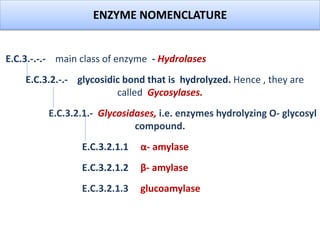
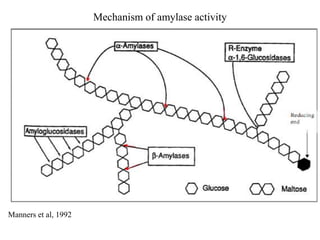
![Various types of amylase associated with degradation of starch and
related polysaccharides structures have been detected and studied[5].
1. Enzymes that hydrolyze 𝛼-1,4 bonds e.g. 𝜶 -amylase (endoacting
amylases).
2. Enzymes that hydrolyze 𝛼 -1,4 e.g. -𝜷 amylase (exoacting amylases
producing maltose as a major end product).
3. Enzymes that hydrolyze terminal 1,4 linked 𝛼 D-glucose residues. e.g.
glucoamylase.
4. Enzymes that hydrolyze only 𝛼 -1,6 linkages e.g. pullulanase .
5. Enzymes that hydrolyze preferentially 𝛼 -1,4 linkages in short chain
oligosaccharides produced by the action of other enzymes on amylose
and amylopectin e.g. - 𝜶 glucosidases.
[5]- Van et al.,2002
TYPES OF AMYLASE](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-5-320.jpg)
![Payan 2004.
The human α-amylase is a classical calcium-containing
enzyme composed of 512 amino acids with a molecular
weight of 57.6 kDa[9]. .
The protein contains 3 domains: A, B, and C.
The A domain (residues 1-99, 169-404 )is the largest,
presenting a typical Tim barrel shaped (β/α)8 super
structure.
The B domain (residues 100-168) is the smallest
domain is attached to the A domain by disulphide
bond. The C domain (residues 405-512) is made up of
anti-parallel beta-structure and is only loosely
associated with Domains A and B.
The active site of the α-amylase is situated in a cleft
located between the carboxyl end of the A and B
domains. Asp206, Glu230 and Asp297 participate in
catalysis [10].
The calcium (Ca2+) is situated at B domain (Asn 100, Arg
158, Asp 167) against the wall of the barrel of Domain
A .
Chloride ion is present at A domain (Arg 195, Asn 298,
and Arg 337)
These ions are required for the stabilization of the
three-dimensional structure .
STRUCTURAL CHARACTERISTICS OF α-AMYLASE
Structure human α-amylase.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-8-320.jpg)
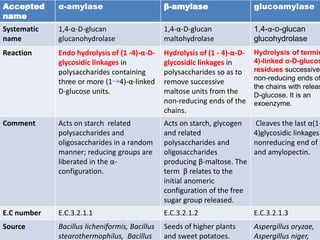
![–
Characterization of α-amylase.
SOURCE Km
(mg
/ml)
Vmax
(μmol/m
g/min)
Kcat
(S-1)
Kcat/
Km
(ml
mg-1
S-1)
Inhibi
tors
Activat
or
Temperat
ure
pH Referen
ces
B.
Licheniformis
6.2 1.04 2000 3.22×
10-2
Hg2+
,Cd2+,
Mn2+,,
Ba2+,
Cu2+,
,EDTA
1500 Da
PEG,
increas
es the
enzyme
activity
by 24%
at
0.02%
w/v
85-90°C 6.5 [11]
Bacillus
megaterium
9.0 0.68 580 .64x
10-2
Hg2+
Ba2+,
,Zn2+,
Co2+,
Cr3+,
Fe3+
Titron X
increase
s the
enzyme
activity
by 34%
at
37-40°C 6.0 [12]](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-9-320.jpg)
![DETERMINATION OF AMYLASE ACTIVITY
• Amylase activity was estimated by measuring either the appearance of one of the
products or the disappearance of the substrate over time.
• The Enzyme – substrate reaction can be determined by measuring the increase in
reducing sugars using the 3, 5 Dinitro salicylic acid reagent[13].
• The pale yellow colored the 3, 5- dinitro salicylic acid undergo reduction in
presence of reducing sugar to yield orange colored 3- amino -5-nitrosalicylic acid.
• The absorbance of resultant solutions is read at 540nm. The intensity of color
depends on the concentration of reducing sugars produced.
Lever et al.,1972.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-10-320.jpg)
![• One unit of amylase activity is defined as the amount of enzyme that
produces 1 μmol of reducing sugar per minute under specific conditions.
Enzyme activity =
U/ml incubation time(min) X volume of starch ( ml)X Volume of cubette
(cubic meter)
• The hydrolytic activity of Amylase can be determined based on the
principle that starch and iodine react to form a blue colored
complex[14].
• On hydrolysis of starch this complex changes. The absorbance can be
read after the enzyme substrate reaction has been terminated.
Conc. Of reducing sugar(µmol) volume content
Obtained from standard graph X in tube X dilution factor](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-11-320.jpg)
![Microorganism Fermentation pH
optimal/stability
Temperature
optimal/stability
Reference
Bacteria
Bacillus
amyloliquefaciens
SmF 7.0 33 °C [16]
Bacillus subtilis SSF 7.0 37 °C [17]
Fungi
Aspergillus niger SSF 5.5 70 °C [18]
Aspergillus
fumigatus
SmF 6.0 30°C [19]
Amylase is ubiquitous enzyme produced by plants, animals and microbes.
In the recent past, there has been extensive research on microbial production of
Amylase.
300 tonnes of α-Amylase have been accounted to be produced from B.lichinoformis
and Aspesgillus sp. per year[15].
There are two methods widely used for production of α-Amylase on a commercially -
1) Submerged fermentation
2) Solid State fermentation](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-12-320.jpg)
![APPLICATION
• Amylases constitute a major class of industrial enzymes which alone form 25% of
the enzyme market covering industrial processes.
Industrial
application
Microbial
source
Role Refere
nce
Starch
conversion
B.amyloliquefaciens,
B.licheniformis
gelatinization, liquefaction,
saccharification of starch
[20]
Bakery Bacillus
stearothermophilus
Converting starch in dough to smaller
fermentable sugars.
[21 ]
Detergent
Industry
Bacillus sp
Aspergillus sp
Degrade the residues of starchy foods
such as potatoes, gravies, custard,
chocolate, etc. to dextrins and other
smaller oligosaccharides .
[ 22]
Textile Industry Bacillus sp Used in removal of starch sizing agent
from woven fabric.
[23 ]
Fuel Production E.coli, B.subtilis Converting starch in to smaller
fermentable sugars which are acted upon
by yeast to produce ethanol.
[24 ]
Paper industry Bacillus sp Viscosity of the natural starch is too high
for paper sizing and this can be altered by
[25]](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-13-320.jpg)
![REFERENCES.
[1] Sivaramakrishnan S., Gangadharan D.,Madhavan K., Ricardo C., Pandey A. α-Amylases from
Microbial Sources. Food Technol. Biotechnol. 2006; 44 (2):173-184.
[2] Aiyer, P.V. Amylase and their applications. African Journal of Biotechnology.2005; 4(13): 1525
- 1529.
[3] Windish, W., Mhatre, N.S. Microbial amylases. Advances in applied microbiology,.2005;7:273
- 304.
[4] Van M., Leemhuis H, Dijkhuizen L.Properties and applications of starch converting enzymes
of the α-amylase family, J. Biotechnol. 2002; 94:137-155.
[5] Manners, D.J. Enzymatic synthesis and degradation of starch and glycogen. Adv. Carbohydr.
Chem. (1992);17:371–430.
[6] Gangadharan, D., Nampoothiri, K. M., Soccol, C. R., & Pandey, A. α-Amylases from Microbial
Sources. Food Technology & Biotechnology.2006.,44(2):23-27.
[7] Kaplan F., Dong S., Charles L. “Roles of β-amylase and starch breakdown” .International
Journal on Plant Physiology. 2006., 126:120–128.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-14-320.jpg)
![[8] Reddy N.S., Nimmagadda A., Sambasiva K.R.S. An overview of the microbial
amylase family. Afr. J. Biotechnol. 2003;2:645–648.
[9] Payan F. Structural basis for the inhibition of mammalian and insect alpha-
amylases by plant protein inhibitors. Biochem Biophys Acta. 2004;1696:171–180.
[10] Muralikrishna G., Nirmala M. Cereal α-amylases—an overview. Carbohydrate
Polymers. 2005;60:163–173.
[11] Saptadip S., Das A., Kumar H., Jana A. Thermodynamic and kinetic
characteristics of an α-amylase from Bacillus licheniformis SKB4. Acta Biol
Szeged.2014; 58(2):147-156 .
[12] Tanaka, A and Hoshino, E. 2002. Calcium binding parameter of Bacillus
amyloliquefaciens amylase determined by inactivation kinetics. Biochemistry
Journal, 364: 635 – 639.
[13] Miller, G.L., Use of dinitrosalicylic acid reagent for determination of reducing
sugar, Anal. Chem.1959; 31:426-428.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-15-320.jpg)
![[14] Hamilton, L. M., Kelly, C. T., & Fogarty, W. M. Carbohydrate Research.1998; 314, 251–257.
[15] Chi Z., Liu G., Wang F., Ju L., Zhang T. Saccharomycopsis fibuligera and its applications in
biotechnology. Biotechnol Adv. 2009;27:423–431.
[16] Gupta R., Gigras P., Mohapatra H., Goswami V.K., Chauhan B. Microbial α-amylases: a
biotechnological perspective. Process Biochem. 2003;38:1599–1616.
[17] Tanyildizi M.S., Ozer D., Elibol M. Production of bacterial α-amylase by B. amyloliquefaciens
under solid substrate fermentation. Biochem. Eng. J. 2007;37:294–297.
[18] Baysal Z., Uyar F., Aytekin C. Solid state fermentation for production of α-amylase by a
thermotolerantBacillus subtilis from hot-spring water. Process Biochemistry. 2003;38:1665–
1668.
[19] Uguru G.C., Akinyauju J.A., Sani A. The use of yam peel for growth of locally
isolated Aspergillus niger and amylase production. Enzyme Microb. Technol. 1997;21:46–51.
[20] Jin B., Leeuwen H.J., Patel B., Yu Q. Utilisation of starch processing wastewater for
production of microbial biomass protein and fungal α-amylase by Aspergillus oryzae. Bioresour.
Technol. 1998;66:201–206.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-16-320.jpg)
![[20] Reddy N.S., Nimmagadda A., Sambasiva Rao K.R.S. An overview of the microbial α-amylase
family. Afr. J. Biotechnol.2003;2:645–648.
[21] Couto S.R., Sanromán M.A. Application of solid-state fermentation to food industry- A
review. Journal of Food Engineering. 2006;76:291–302.
[22] Hmidet N., El-Hadj Ali N., Haddar A., Kanoun S., Alya S., Nasri M. Alkaline proteases and
thermostable α-amylase co-produced by Bacillus licheniformis NH1: Characterization and
potential application as detergent additive. Biochemical Engineering Journal. 2009;47:71–79.
[23] Feitkenhauer H. Anaerobic digestion of desizing wastewater: influence of pretreatment and
anionic surfactant on degradation and intermediate accumulation. Enzyme Microb. Technol.
2003;33:250–258.
[24] Moraes L.M.P., Filho S.A., Ulhoa C.J. Purification and some properties of an α-amylase
glucoamylase fusion protein from Saccharomyces cerevisiae. World J. Microbiol. Biotechnol.
1999;15:561–564.
[25] Bruinenberg P.M., Hulst A.C., Faber A., Voogd R.H. A process for surface sizing or coating of
paper. In: European Patent Application. 1996.](https://image.slidesharecdn.com/enzymologyppt-160824085742/85/Presentation-on-Amylase-enzyme-17-320.jpg)
