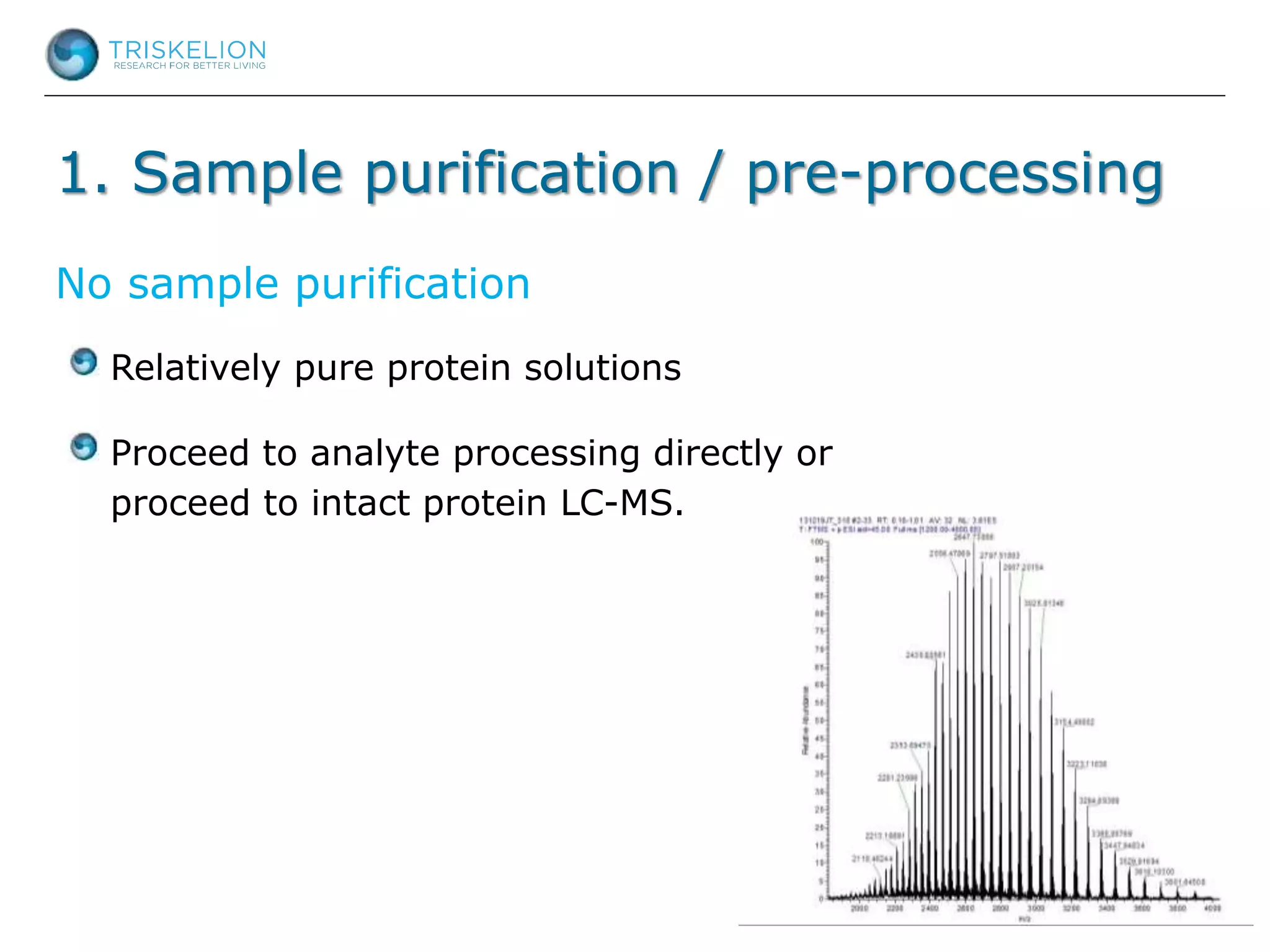
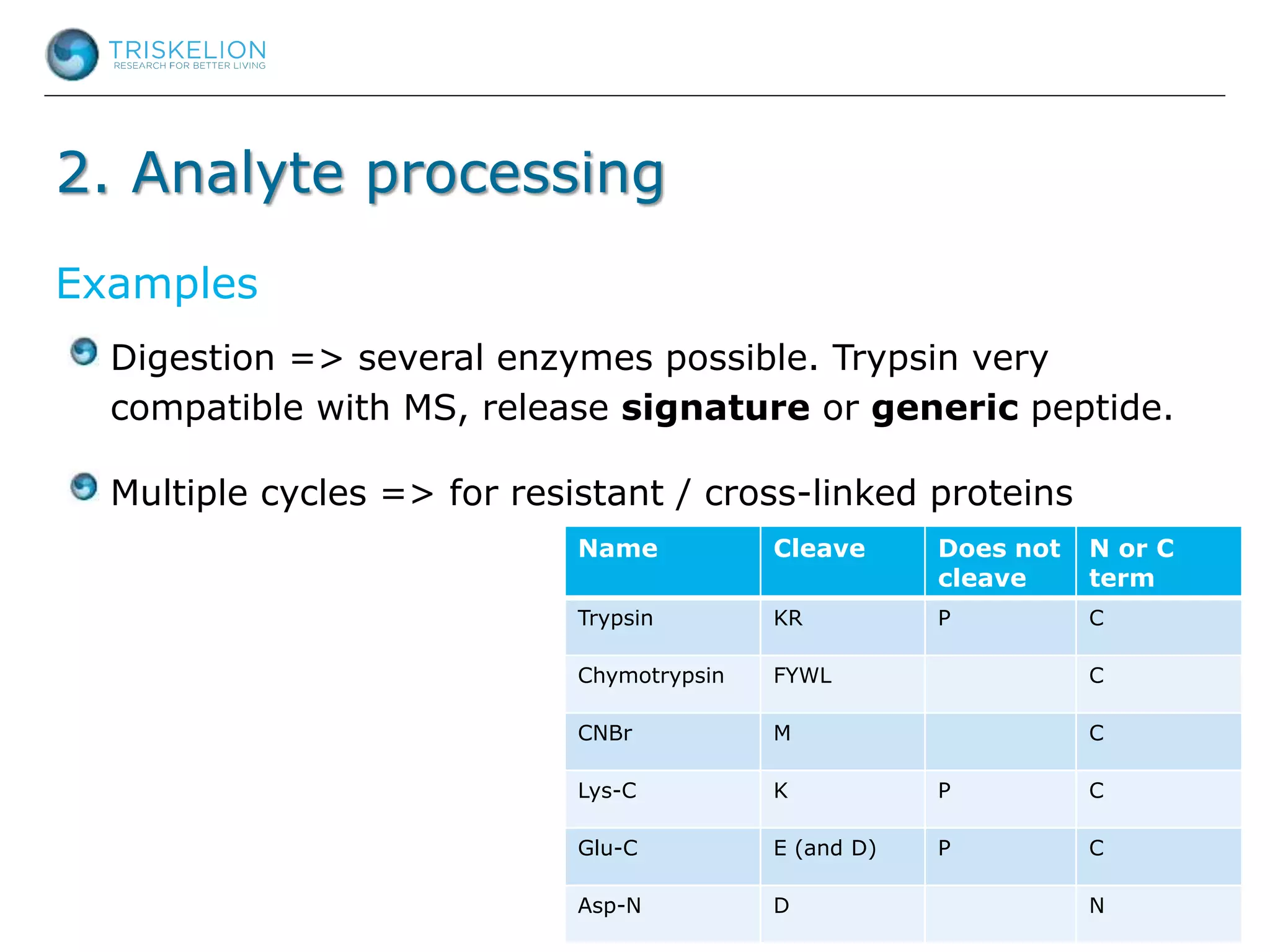
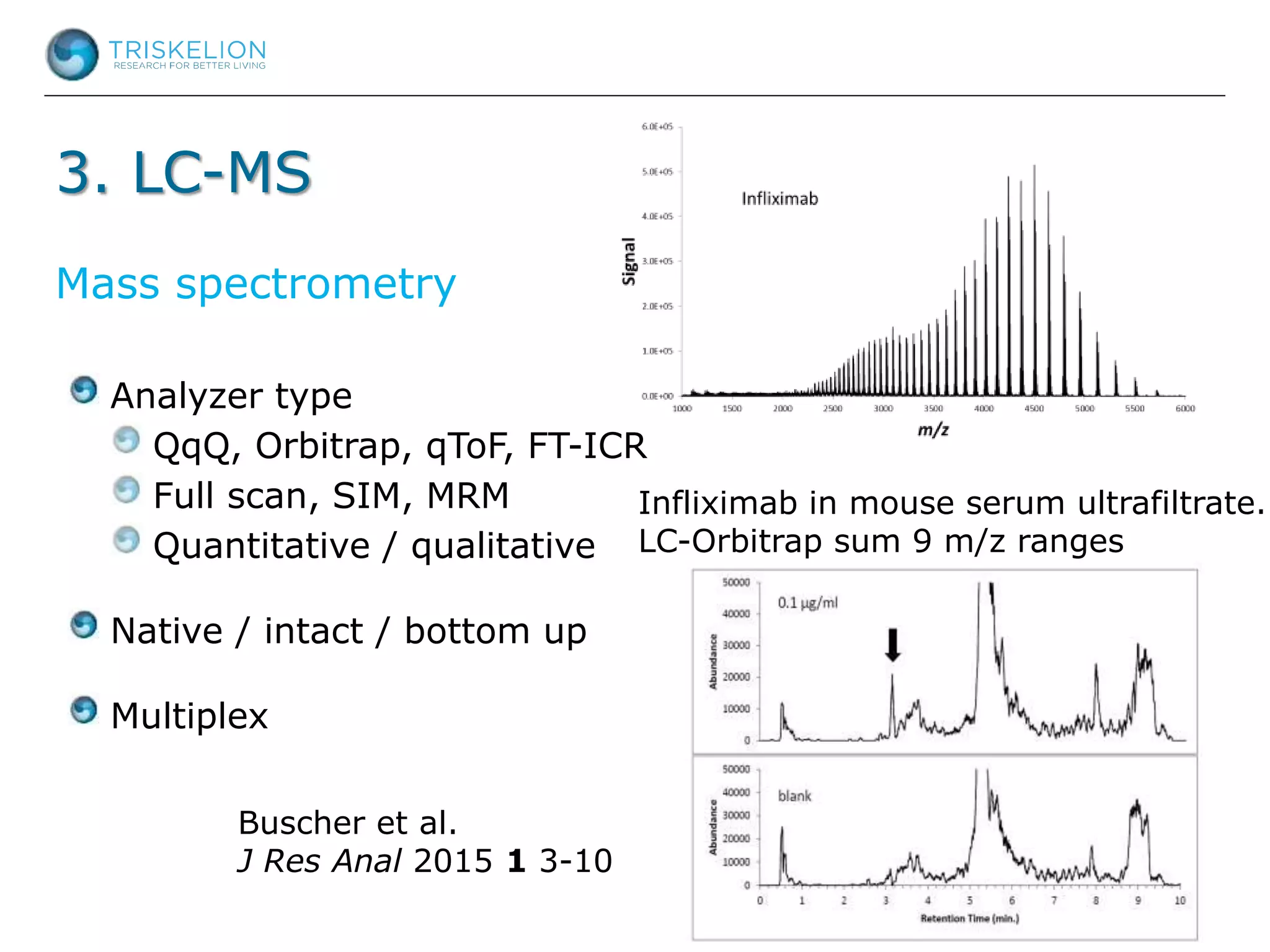
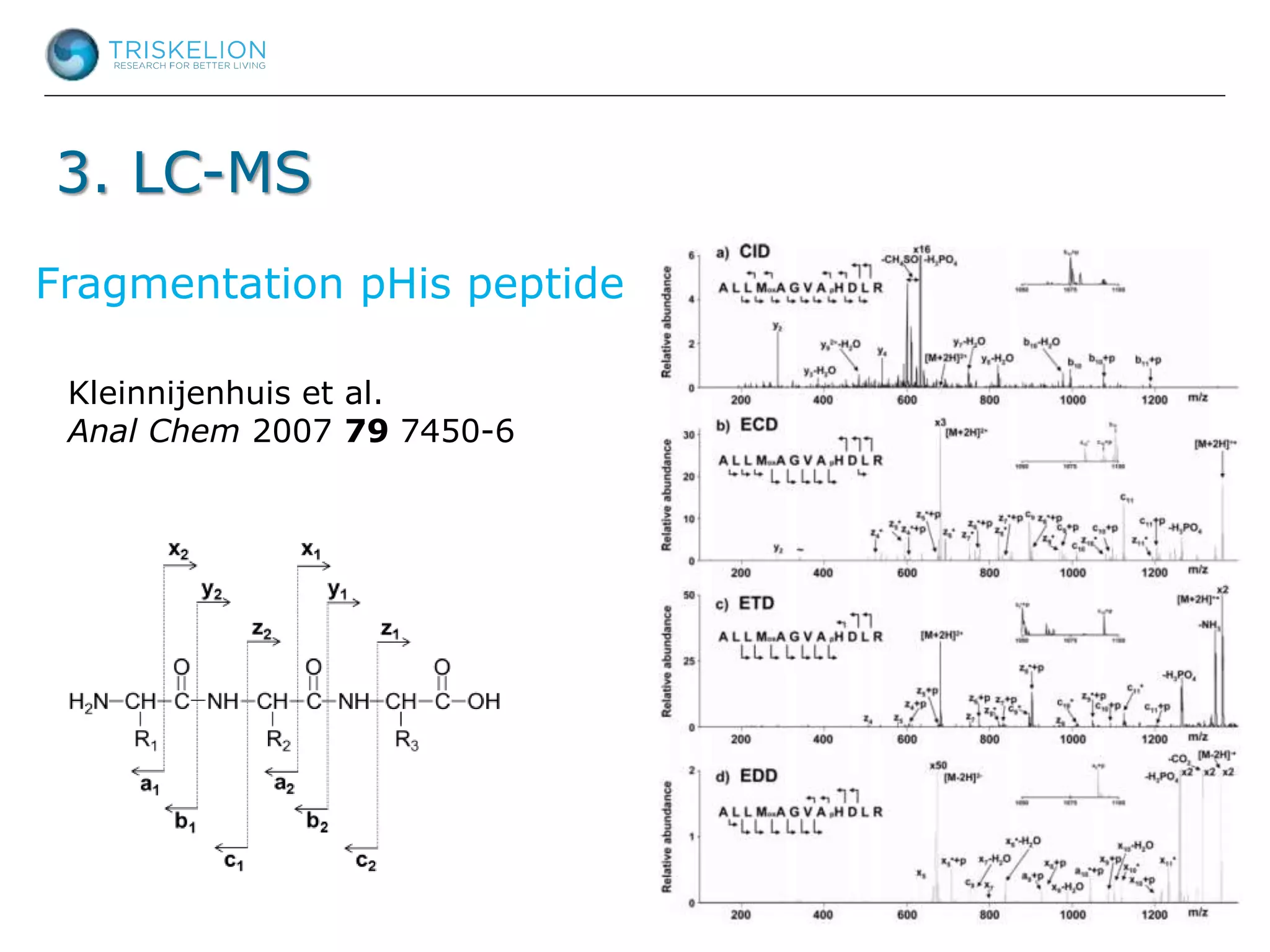
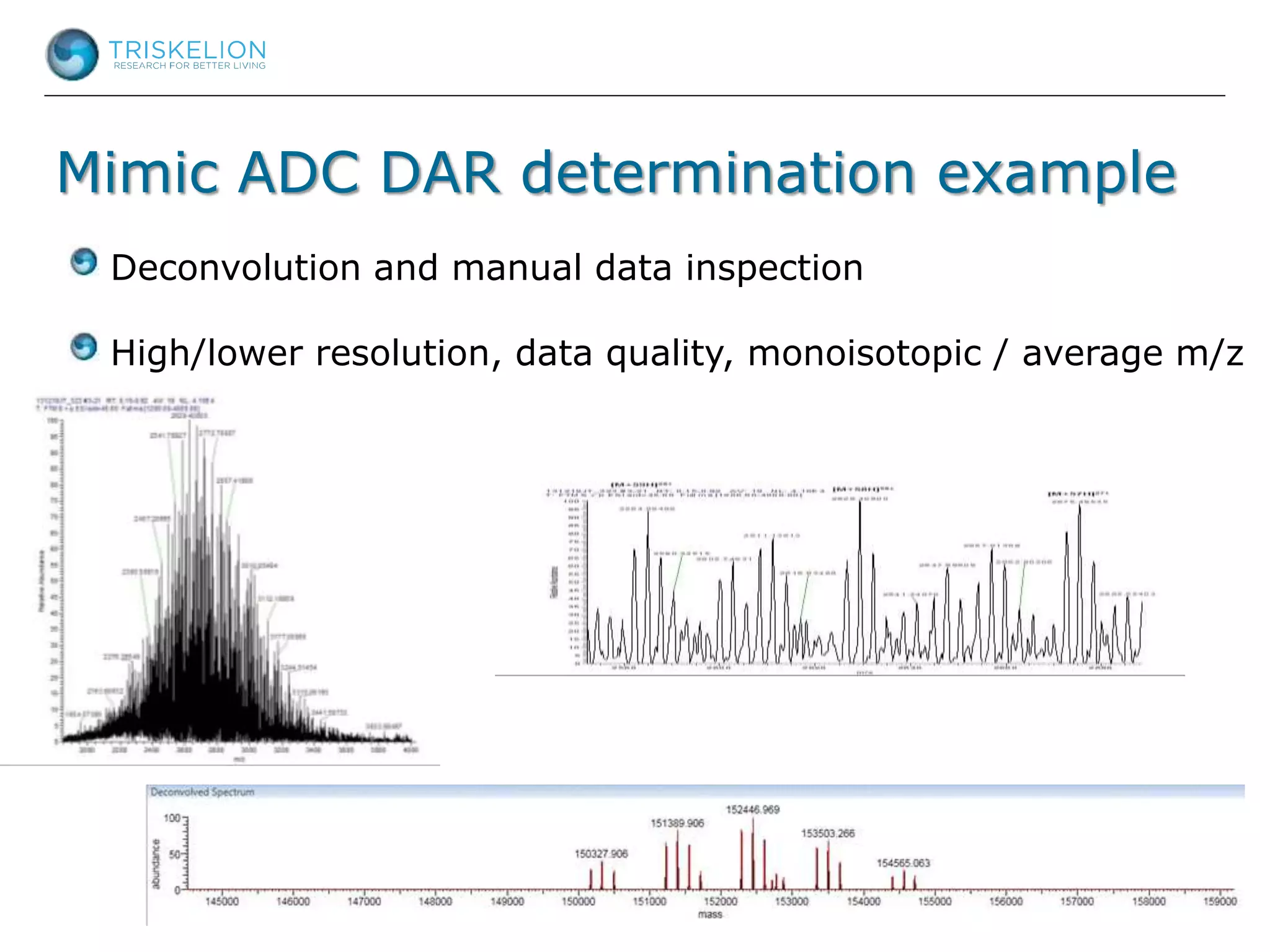
The document outlines strategies for bioanalysis of proteins using liquid chromatography-mass spectrometry (LC-MS), highlighting key steps such as sample purification, analyte processing, and the specifics of LC-MS methods. It details various purification techniques, analyte processing steps, and LC-MS fragmentation methods to enhance detection and analysis of proteins and their characteristics. Additionally, it provides examples and considerations for target selection and sample requirements in LC-MS workflows.


![Triskelion general experimental set up
Absolute recovery
𝐶𝐸𝑖𝑠 =
𝑉𝐿 𝑖𝑠
𝑉𝐸 𝑖𝑠
𝐶𝐿𝑖𝑠𝐶𝐸 𝑝𝑒𝑝 =
𝑀𝑊𝑝𝑒𝑝 × 2
𝑀𝑊𝑝𝑟𝑜𝑡
𝑉𝐿 𝑝𝑟𝑜𝑡
𝑉𝐸 𝑝𝑟𝑜𝑡
𝐶𝐿 𝑝𝑟𝑜𝑡
𝑦 =
𝐶𝐸 𝑝𝑒𝑝
𝐶𝐸𝑖𝑠
𝑦 = 𝑎𝑥 + 𝑏
Corrections isotopic
disturbance / molar
𝑎 =
2 × 𝑉𝐿 𝑝𝑟𝑜𝑡 × 𝑀𝑊𝑖𝑠
𝑀𝑊𝑝𝑟𝑜𝑡 × 𝑉𝐿𝑖𝑠 × 𝐶𝐿𝑖𝑠
𝐹1 𝑝𝑒𝑝
𝐹1 𝑖𝑠
DILLTQSPAILSVSPGER
DILLTQSPAILSVSPGE[R_13C6
15N4]
[M+3H]3+
633.0 634.0 635.0 636.0 637.0 638.0m/z
0
50
100
RelativeAbundance
636.024
636.357
633.022632.689
636.690633.356
633.690 637.024
635.691 637.358634.024
𝑥 = 𝐶𝐿 𝑝𝑟𝑜𝑡](https://image.slidesharecdn.com/annekleinnijenhuis-160523191710/75/Strategies-for-bioanalysis-of-proteins-using-LC-MS-28-2048.jpg)