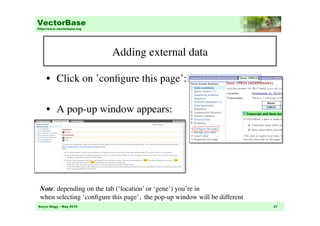
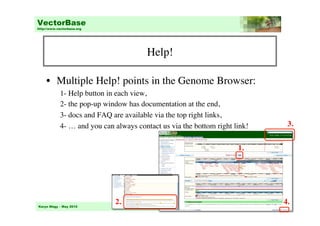
This document provides an overview of genome browsing at VectorBase, an online genomic database for invertebrate vectors of human pathogens. It describes the species available, data types, and how to navigate the main page and genome browser. Key features of the genome browser include viewing genomic regions at different levels of detail, searching or browsing to genes of interest, and adding custom tracks with additional data. Help documentation is also available.