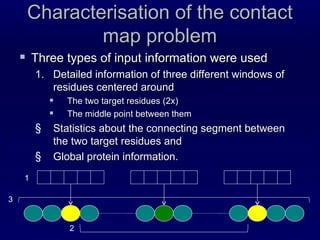
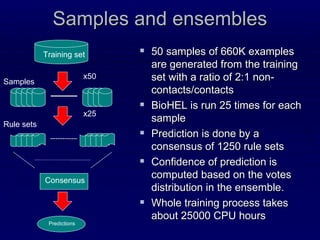
The document describes the Infobiotics Contact Map Predictor submitted to CASP9. It summarizes the steps taken to predict protein contact maps, which include predicting secondary structure, solvent accessibility, recursive convex hull, and coordination number. These predictions are integrated using BioHEL and based on local sequence and evolutionary information within windows of neighboring residues. The dataset used for training and testing contained over 32 million instances from 3262 protein chains with resolutions under 2 Angstroms and less than 30% sequence identity.

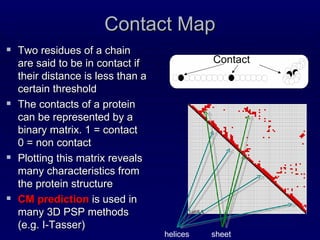
![Steps Prediction of Secondary structure (using PSIPRED) Solvent Accessibility Recursive Convex Hull Coordination number Integration of all these predictions plus other sources of information Final CM prediction (using BioHEL) Using BioHEL [Bacardit et al., 09]](https://image.slidesharecdn.com/infobioticscmpredictor-110531110854-phpapp02/85/The-Infobiotics-Contact-Map-predictor-at-CASP9-3-320.jpg)