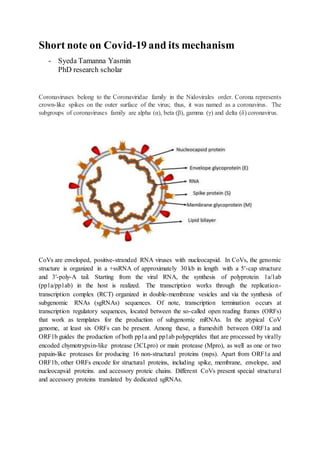
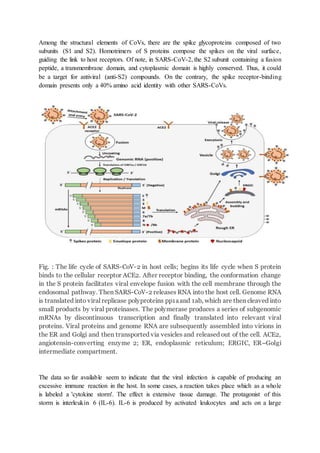
Coronaviruses are enveloped viruses with positive-stranded RNA genomes that encode structural and accessory proteins. SARS-CoV-2 enters cells by binding to the ACE2 receptor, and its RNA genome is then translated and replicated to produce more viral proteins and RNA. Assembly and release of new virus particles occurs in the ER and Golgi. The virus can trigger a cytokine storm involving overproduction of IL-6 that may lead to severe immune response and multi-organ dysfunction in some patients.