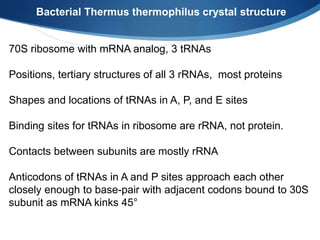
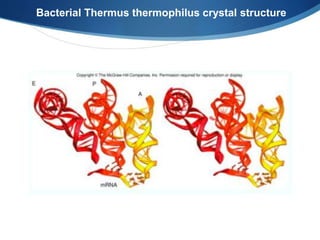
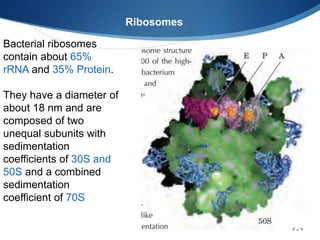
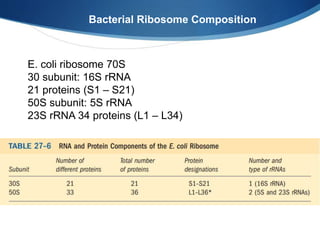
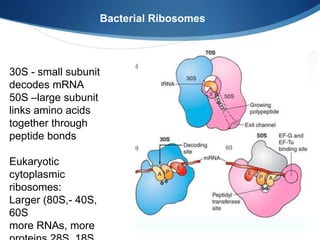
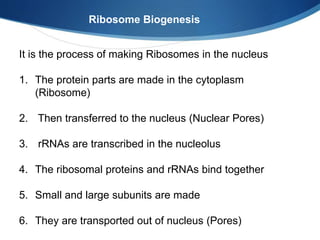
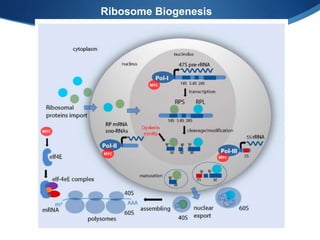
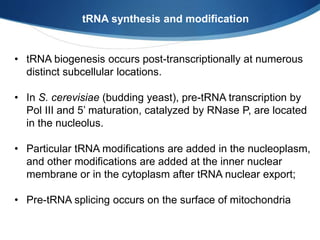
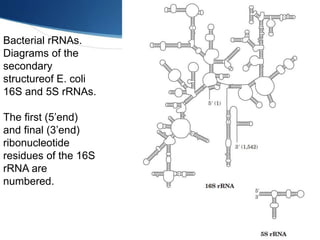
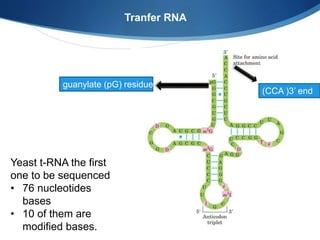
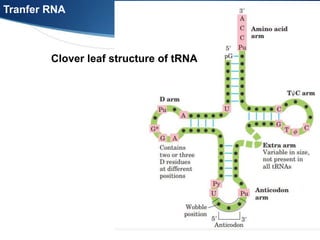
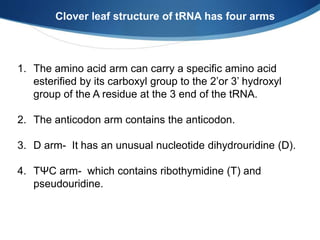
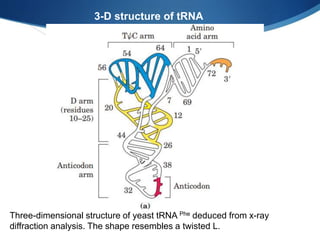
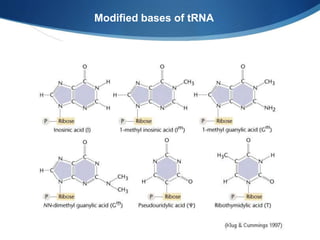
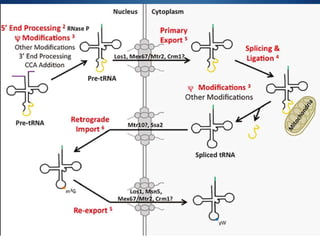
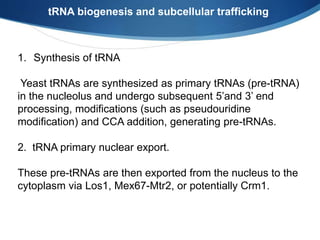
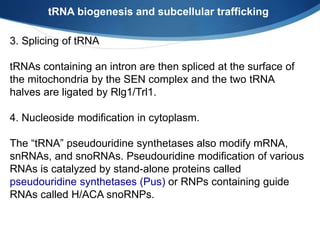
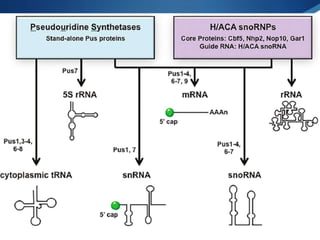
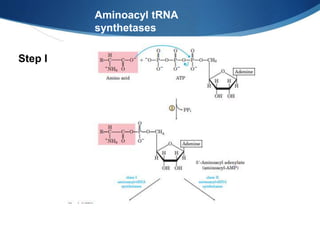
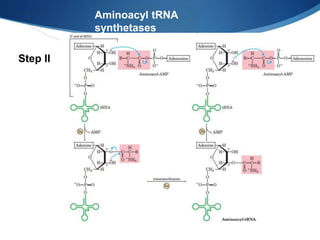
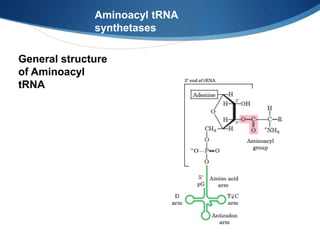
Ribosomes are structures composed of rRNA and proteins that synthesize proteins. They consist of a small 30S subunit and large 50S subunit. The 30S subunit decodes mRNA and the 50S subunit links amino acids. Transfer RNA has a cloverleaf structure and serves as an adapter between mRNA and amino acids. Important discoveries included the identification of ribosomes by Palade in 1955 and determination of the ribosome structure including rRNA and protein components by Ramakrishnan, Steitz, and Yonath in 2000.