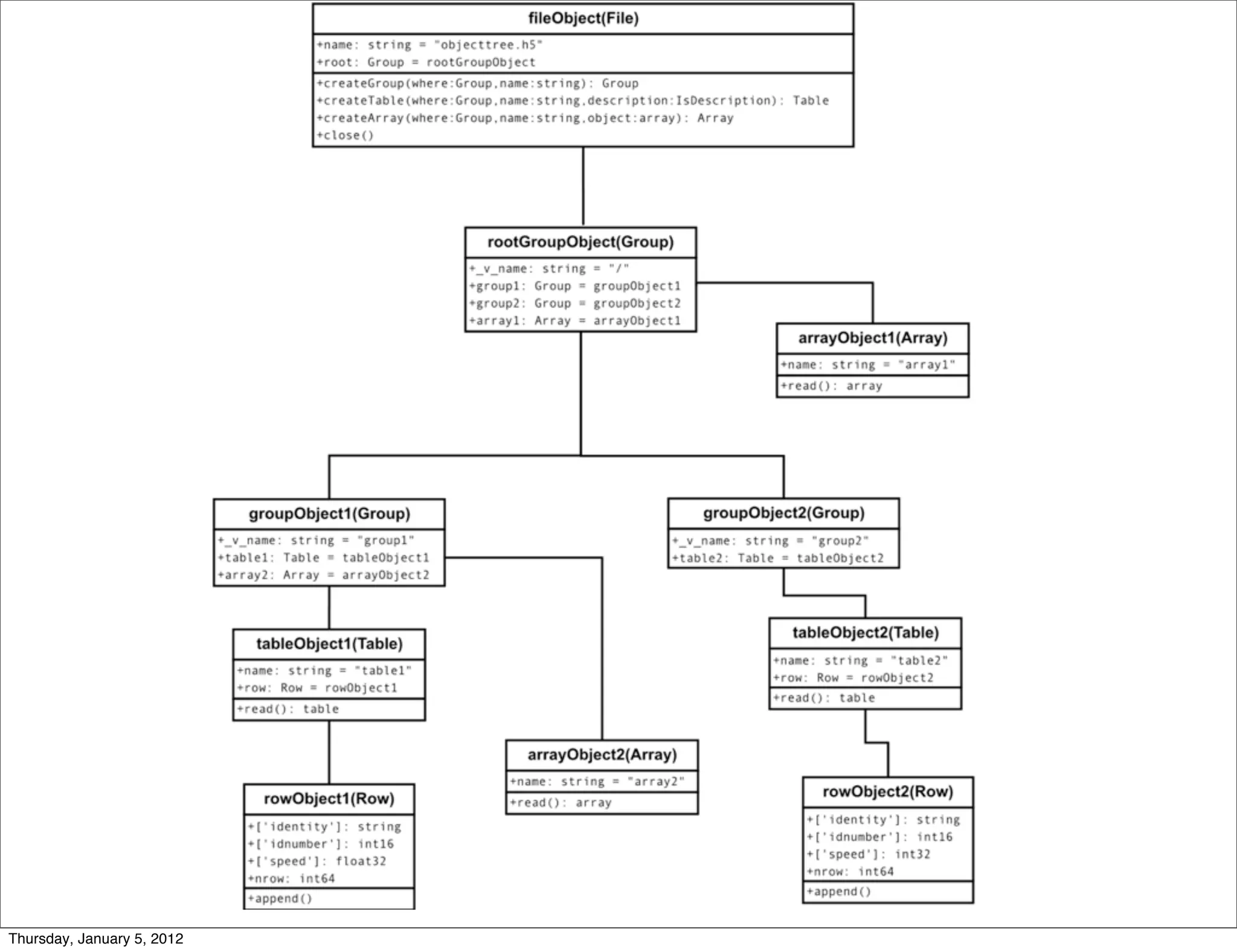
HDF5 is a file format and software library for storing and managing large amounts of numerical data. It supports hierarchical organization of data through groups, datasets that store multidimensional arrays of data, and attributes that store metadata. HDF5 files can be accessed and extended with additional data through its API, allowing for efficient input/output and access to subsets of large datasets.


![from tables import *
# Define a user record to characterize some kind of particles
class Particle(IsDescription):
name = StringCol(16) # 16-character String
idnumber = Int64Col() # Signed 64-bit integer
ADCcount = UInt16Col() # Unsigned short integer
TDCcount = UInt8Col() # unsigned byte
grid_i = Int32Col() # integer
grid_j = Int32Col() # integer
pressure = Float32Col() # float (single-precision)
energy = FloatCol() # double (double-precision)
filename = "test.h5"
# Open a file in "w"rite mode
h5file = openFile(filename, mode = "w", title = "Test file")
# Create a new group under "/" (root)
group = h5file.createGroup("/", 'detector', 'Detector information')
# Create one table on it
table = h5file.createTable(group, 'readout', Particle, "Readout example")
# Fill the table with 10 particles
particle = table.row
for i in xrange(10):
particle['name'] = 'Particle: %6d' % (i)
particle['TDCcount'] = i % 256
particle['ADCcount'] = (i * 256) % (1 << 16)
particle['grid_i'] = i
particle['grid_j'] = 10 - i
particle['pressure'] = float(i*i)
particle['energy'] = float(particle['pressure'] ** 4)
particle['idnumber'] = i * (2 ** 34)
# Insert a new particle record
particle.append()
# Close (and flush) the file
h5file.close()
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-3-2048.jpg)
![Filling a table
>>> class Particle(IsDescription):
... name = StringCol(16) # 16-character String
... idnumber = Int64Col() # Signed 64-bit integer
... ADCcount = UInt16Col() # Unsigned short integer
... TDCcount = UInt8Col() # unsigned byte
... grid_i = Int32Col() # 32-bit integer
... grid_j = Int32Col() # 32-bit integer
... pressure = Float32Col() # float (single-precision)
... energy = Float64Col() # double (double-precision)
>>> table = h5file.root.detector.readout
>>> particle = table.row
>>> for i in xrange(10, 15):
... particle['name'] = 'Particle: %6d' % (i)
... particle['TDCcount'] = i % 256
... particle['ADCcount'] = (i * 256) % (1 << 16)
... particle['grid_i'] = i
... particle['grid_j'] = 10 - i
... particle['pressure'] = float(i*i)
... particle['energy'] = float(particle['pressure'] ** 4)
... particle['idnumber'] = i * (2 ** 34)
... particle.append()
>>> table.flush()
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-5-2048.jpg)
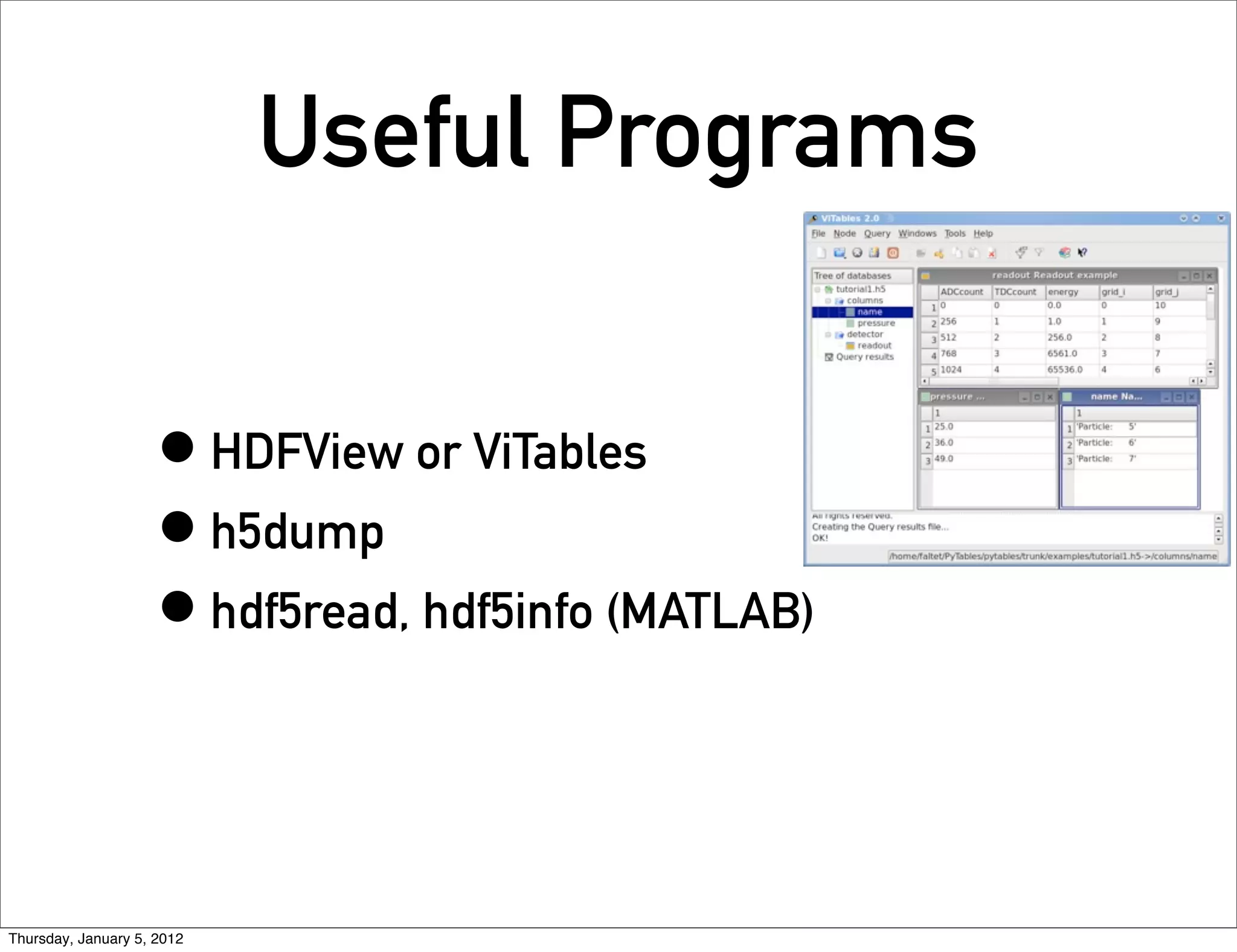
![Accessing a table:
Slicing
>>> table.cols.TDCcount[0] = 1
>>> table.cols.energy[1:9:3] = [2,3,4]
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-6-2048.jpg)
![Search in Tables
>>> class Particle(IsDescription):
... name = StringCol(16) # 16-character String
... idnumber = Int64Col() # Signed 64-bit integer
... ADCcount = UInt16Col() # Unsigned short integer
... TDCcount = UInt8Col() # unsigned byte
... grid_i = Int32Col() # 32-bit integer
... grid_j = Int32Col() # 32-bit integer
... pressure = Float32Col() # float (single-precision)
... energy = Float64Col() # double (double-precision)
>>> table = h5file.root.detector.readout
>>> pressure = [x['pressure'] for x in table.iterrows() if x['TDCcount'] > 3 and 20 <= x
['pressure'] < 50]
>>> pressure
[25.0, 36.0, 49.0]
“In-Kernel” Version
>>> names = [ x['name'] for x in table.where("""(TDCcount > 3) & (20 <= pressure) & (pressure < 50)"
>>> names
['Particle: 5', 'Particle: 6', 'Particle: 7']
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-7-2048.jpg)

![(C)Arrays
import numpy
import tables
fileName = 'carray1.h5'
shape = (200, 300)
atom = tables.UInt8Atom()
filters = tables.Filters(complevel=5, complib='zlib')
h5f = tables.openFile(fileName, 'w')
ca = h5f.createCArray(h5f.root, 'carray', atom, shape, filters=filters)
# Fill a hyperslab in ``ca``.
ca[10:60, 20:70] = numpy.ones((50, 50))
h5f.close()
# Re-open and read another hyperslab
h5f = tables.openFile(fileName)
print h5f
print h5f.root.carray[8:12, 18:22]
h5f.close()
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-9-2048.jpg)
![(E)Arrays
import tables
import numpy
fileh = tables.openFile('earray1.h5', mode='w')
a = tables.StringAtom(itemsize=8)
# Use ''a'' as the object type for the enlargeable array.
array_c = fileh.createEArray(fileh.root, 'array_c', a, (0,), "Chars")
array_c.append(numpy.array(['a'*2, 'b'*4], dtype='S8'))
array_c.append(numpy.array(['a'*6, 'b'*8, 'c'*10], dtype='S8'))
# Read the string ''EArray'' we have created on disk.
for s in array_c:
print 'array_c[%s] => %r' % (array_c.nrow, s)
# Close the file.
fileh.close()
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-10-2048.jpg)

![def _get_pgroup(self, file, p, proj = None):
"""
Get group node of tables.File corresponding to property p.
Creates group node, if it does not exist yet.
:param tables.File file: Handle to HDF5 file to which records are saved.
:param string p: To be recorded property.
:param Projection proj: Projection from which property p is recorded.
:return: Group node corresponding to property p.
"""
SDict = self.sim.config.ShapeDispatch
if not proj:
name = self.sheet.name
else:
name = proj.name
try:
pgroup = file.getNode('/%s_%s' % (p, name,))
except NoSuchNodeError:
pgroup = file.createGroup('/', '%s_%s' % (p, name,))
file.createEArray(pgroup, 'data', Float64Atom(),
flatten((0, SDict[p])))
file.createEArray(pgroup, 'step', Int32Atom(), (0, 1))
return pgroup
def _write_attr(self, pgroup, data):
"""
Helper fn writing provided data and step count to group node (of
tables.File)
:param tables.group.Group pgroup: Group node to which data is saved.
:param numpy.Array data: Data matrix to be recorded.
"""
pgroup.data.append([data])
pgroup.step.append([[self.count]])
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-12-2048.jpg)
![def function(self):
"""
Stores activity submatrices from recordings file per node to 3D array
and returns reshaped 2D version of it.
"""
x = self.x
y = self.y
size = self.size
nnames = self.nnames
array = np.zeros((len(nnames), size, size))
with openFile(self.path, 'r') as file:
for i, nname in enumerate(nnames):
node = file.getNode(nname)
array[i, :, :] =
node.data.read(self.cnt)[0, x : x + size, y : y + size]
return array.reshape(size, size * len(nnames))
Thursday, January 5, 2012](https://image.slidesharecdn.com/pytables-120112035612-phpapp02/75/Pytables-13-2048.jpg)