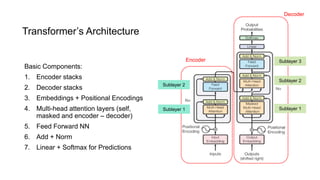
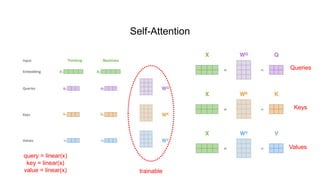
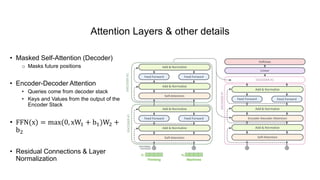
1) The document discusses using a Transformer model for protein secondary structure prediction (PSSP). Transformers use self-attention and avoid issues with long-term dependencies that RNNs face.
2) Several experiments were conducted to determine the best Transformer architecture for PSSP, varying number of layers (N), hidden sizes (d_model), attention heads (h), and other hyperparameters. The best results were around 63-64% accuracy with N=2, d_model=128-256, and h=8.
3) Future work proposed includes exploring n-gram representations, pretraining on larger datasets, and ensembling with other models.