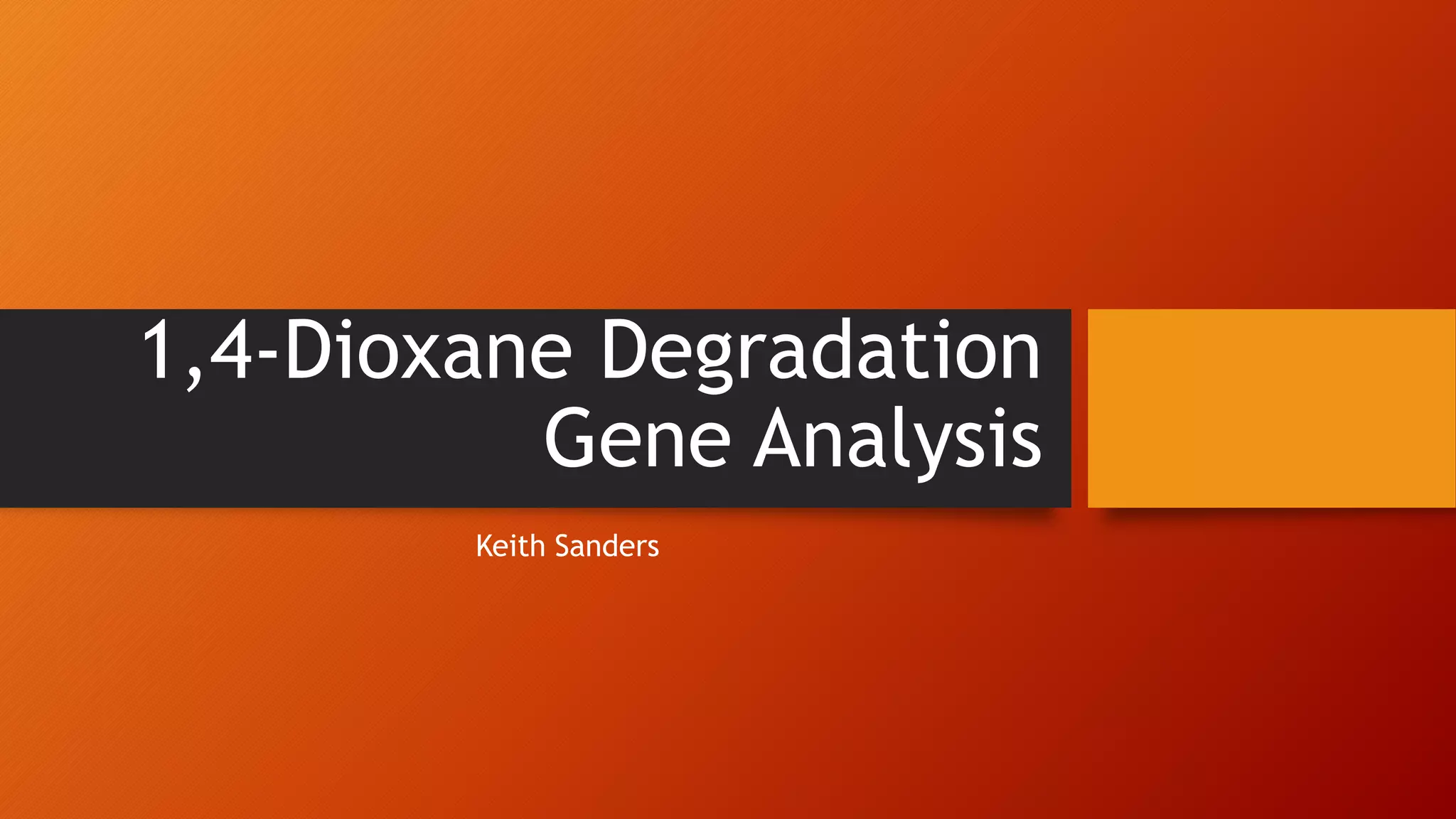
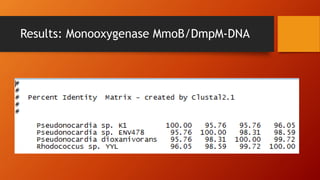
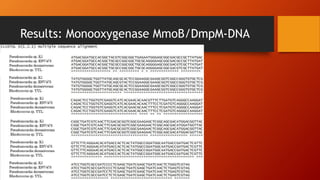
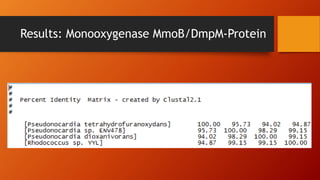
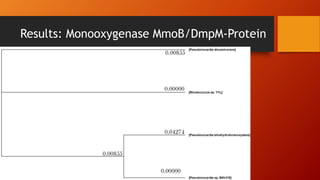
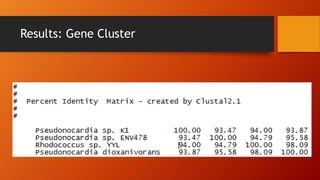
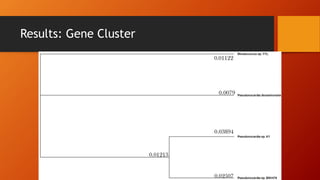
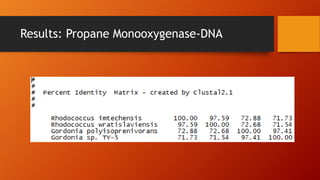
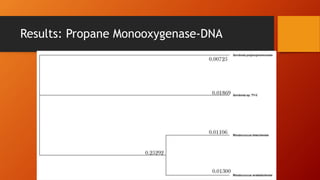
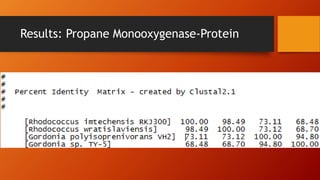
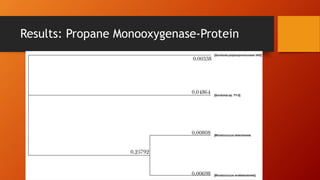
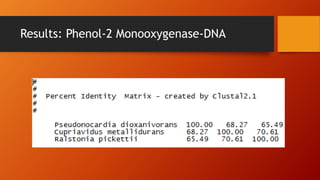
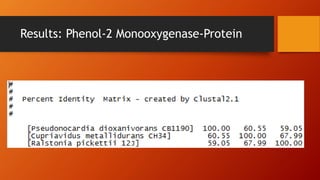
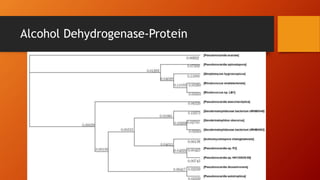
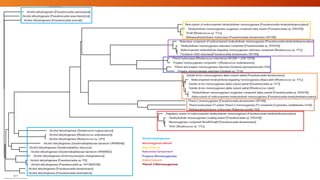
The document summarizes Keith Sanders' research project analyzing genes capable of degrading the solvent 1,4-dioxane. Through literature reviews and database searches, Sanders identified the Monooxygenase MmoB/DmpM gene in Pseudonocardia Dioxanivorans as promising for degradation. Sequence analysis of this gene and related ones in other organisms supported Pseudonocardia sp. K1, Pseudonocardia sp. ENV478, and Rhodococcus sp. YYL as likely capable of true 1,4-dioxane degradation. While other genes may also degrade it, MmoB/DmpM appears most optimal.