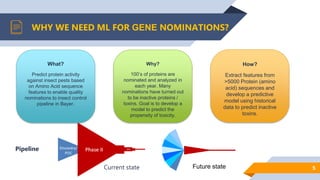
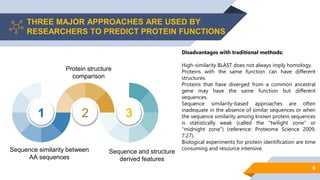
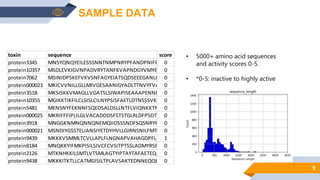
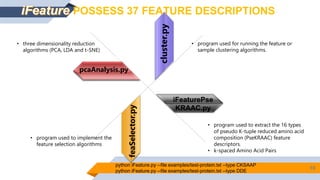
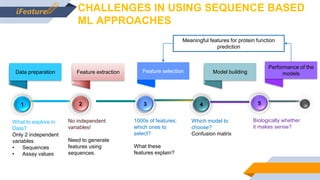
Bayer is utilizing machine learning to enhance the identification of insecticidal proteins to improve crop yields, as traditional methods are inefficient, leading to many inactive nominations. The process involves generating features from over 5000 amino acid sequences using the iFeature toolkit and building predictive models to streamline protein testing for efficacy against pests. The presentation discusses the challenges faced, methods to overcome them, and the importance of domain knowledge in model selection.