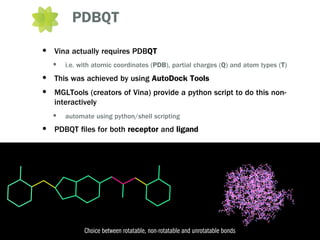
This document outlines the development of OpenDiscovery, an open source platform for automated docking of ligands to proteins and molecular simulation. It describes using freely available tools like AutoDock Vina, OBabel and PyMol to screen a library of compounds against a target protein, generate similar compounds, prepare ligand and receptor files, and visualize and summarize results. Issues around parameterizing novel ligands for molecular dynamics simulation are also discussed. The goal is to create a high-throughput virtual screening workflow to propose ligands for further testing, and contribute to OpenDiscovery which will integrate cheminformatics and additional analysis tools.




c(O)c1)C(=O)O
5
NH2
HO
HO
O
OH](https://image.slidesharecdn.com/mp1-presentation-140608185918-phpapp02/85/OpenDiscovery-5-320.jpg)