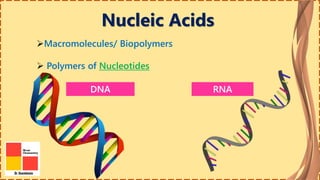
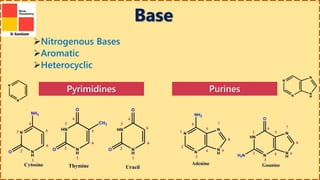
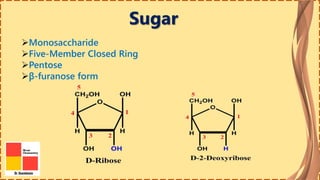
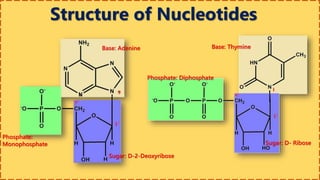
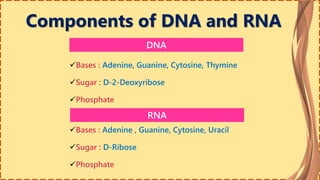
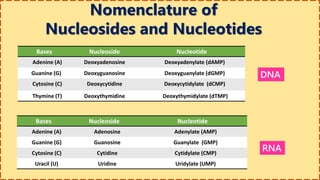
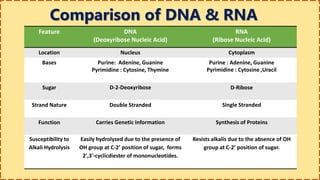
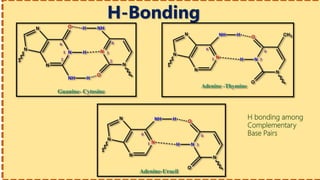
This document provides information on nucleic acids and their components. It discusses that nucleic acids are macromolecules composed of nucleotides that are polymers of either RNA or DNA. The key components of nucleotides are nitrogenous bases, the pentose sugar ribose or deoxyribose, and a phosphate group. Nucleotides serve as monomers that bond together to form nucleic acid polymers through phosphodiester linkages between the 3' carbon of one sugar and the 5' carbon of the next. The document compares the key differences between DNA and RNA such as their bases, sugars, and functions.