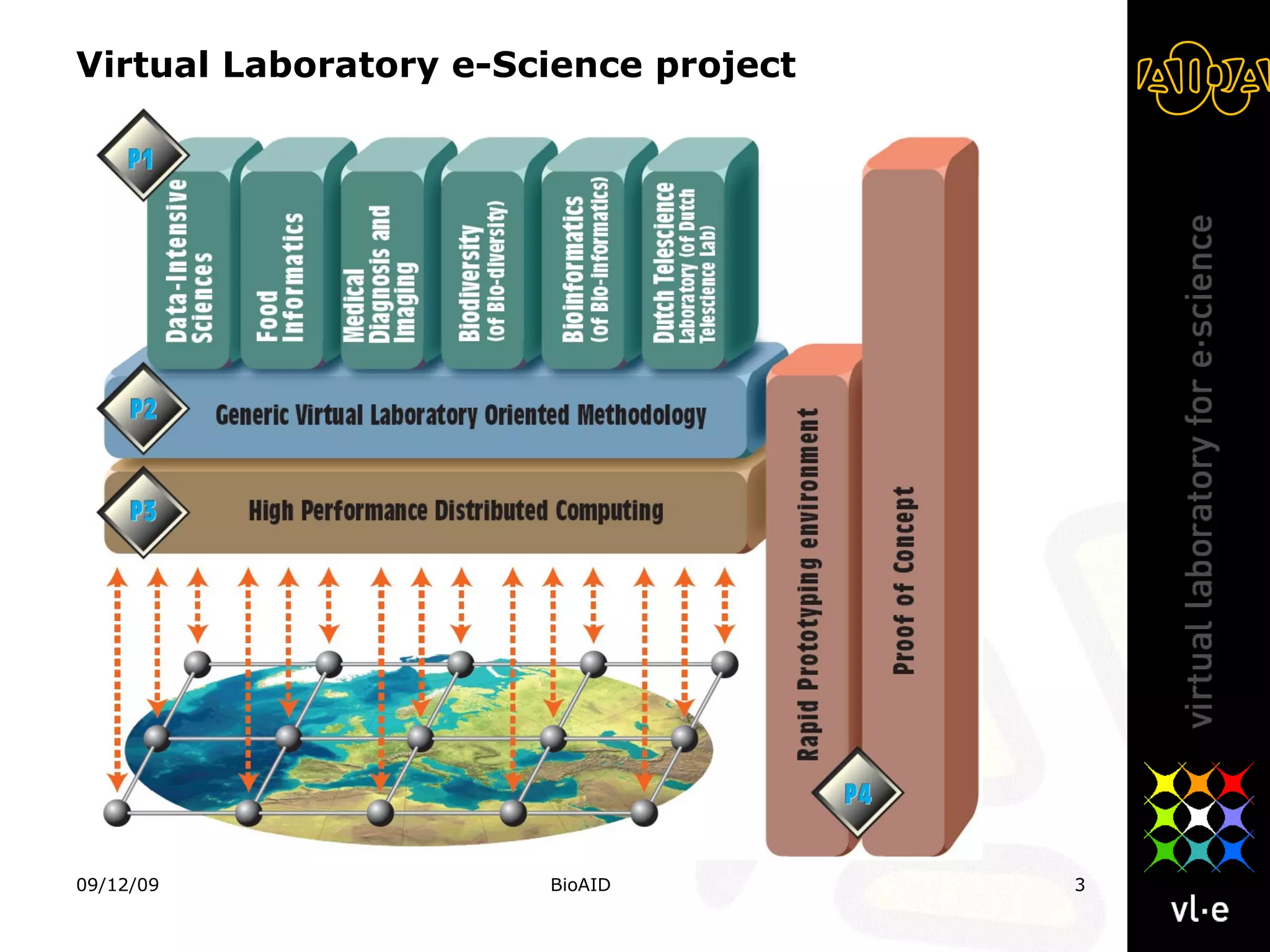
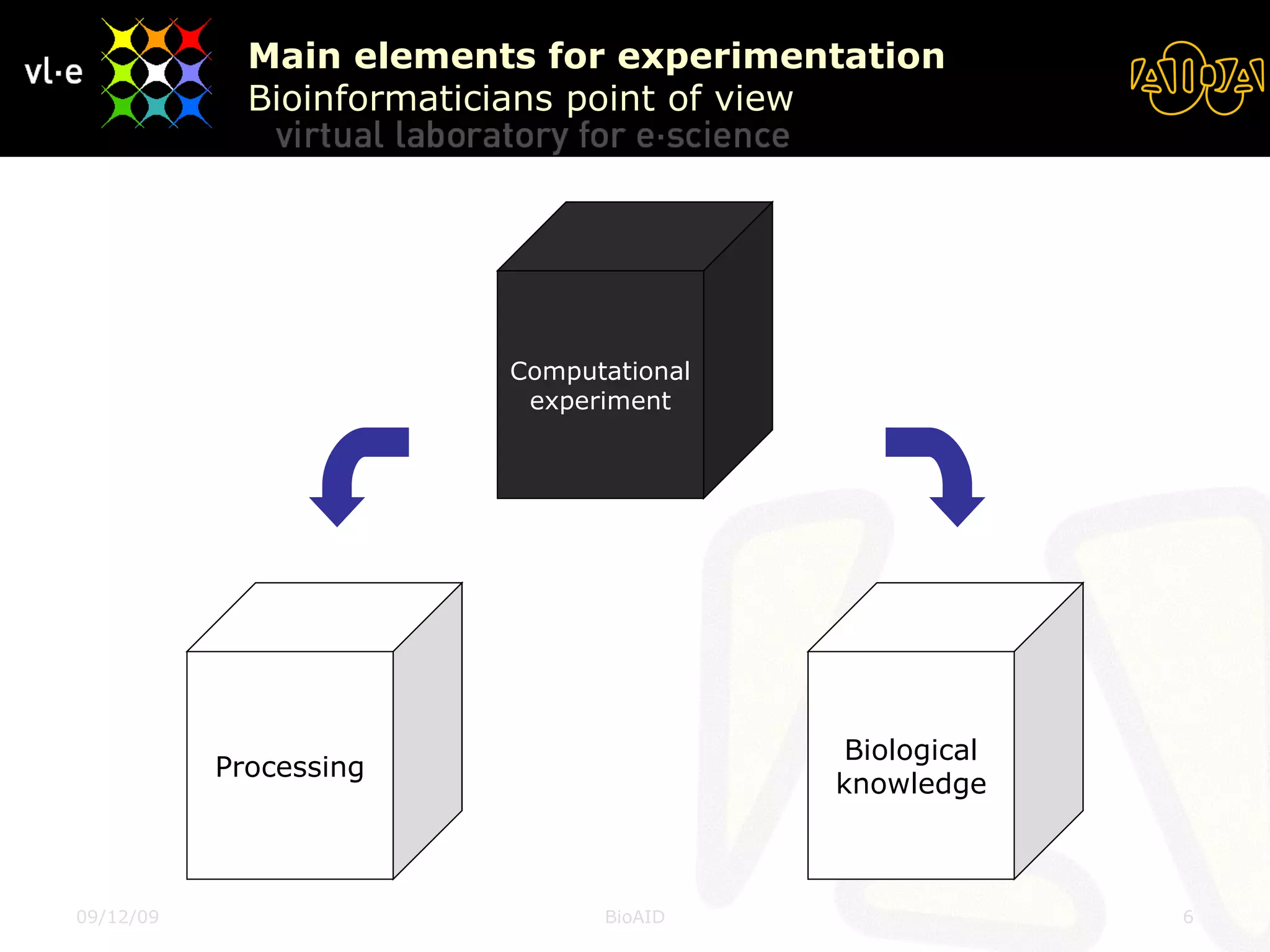
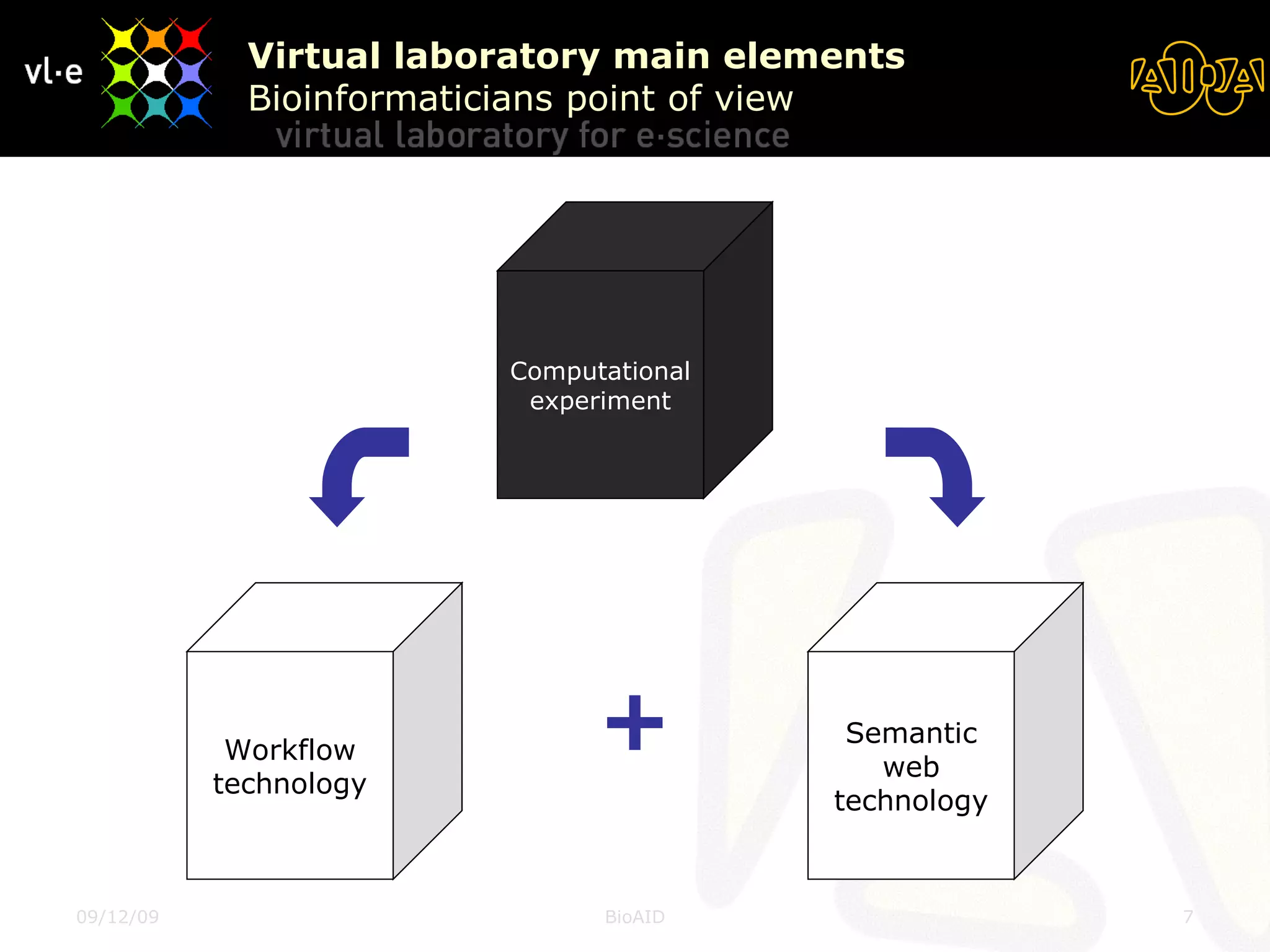
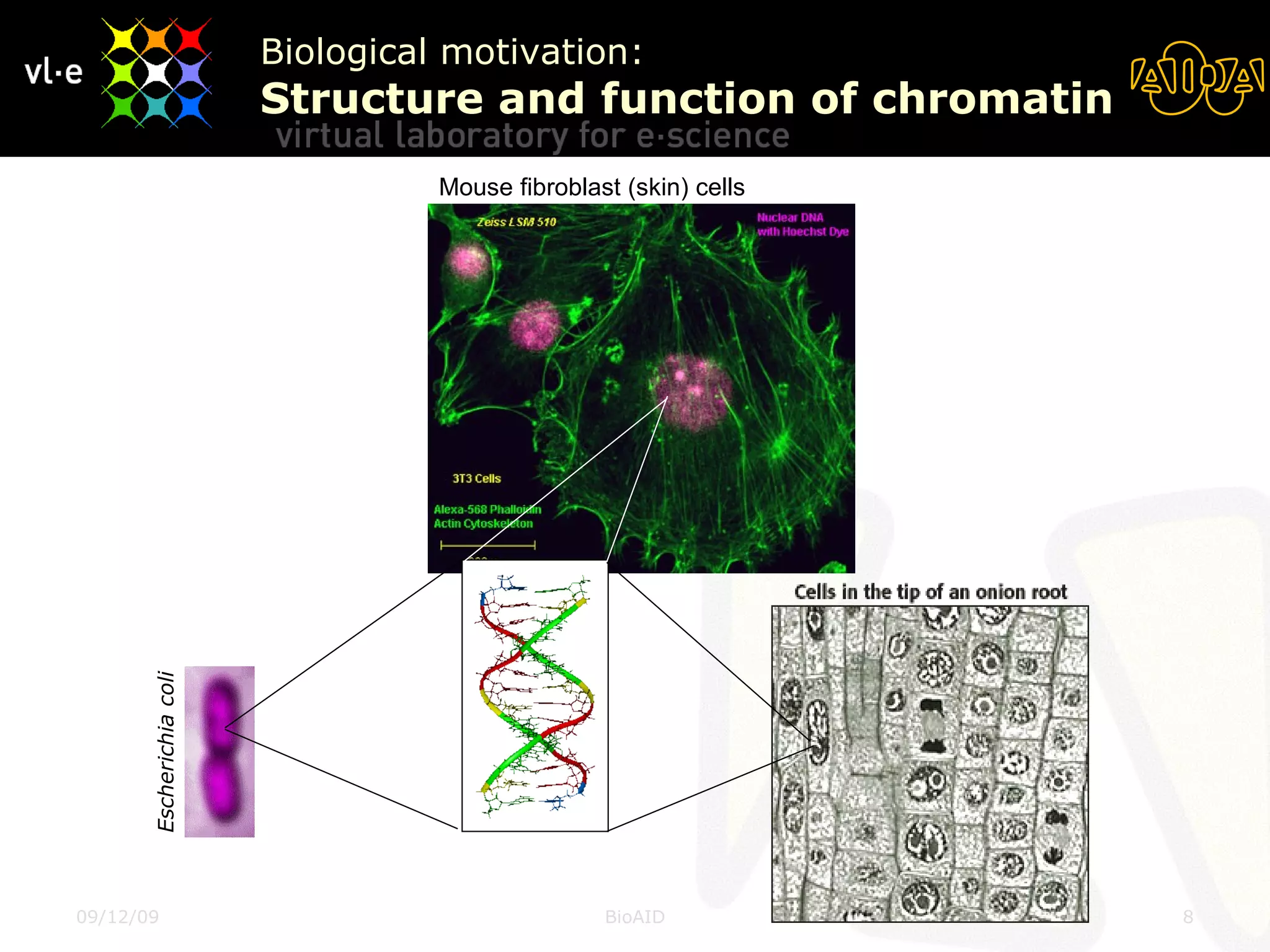
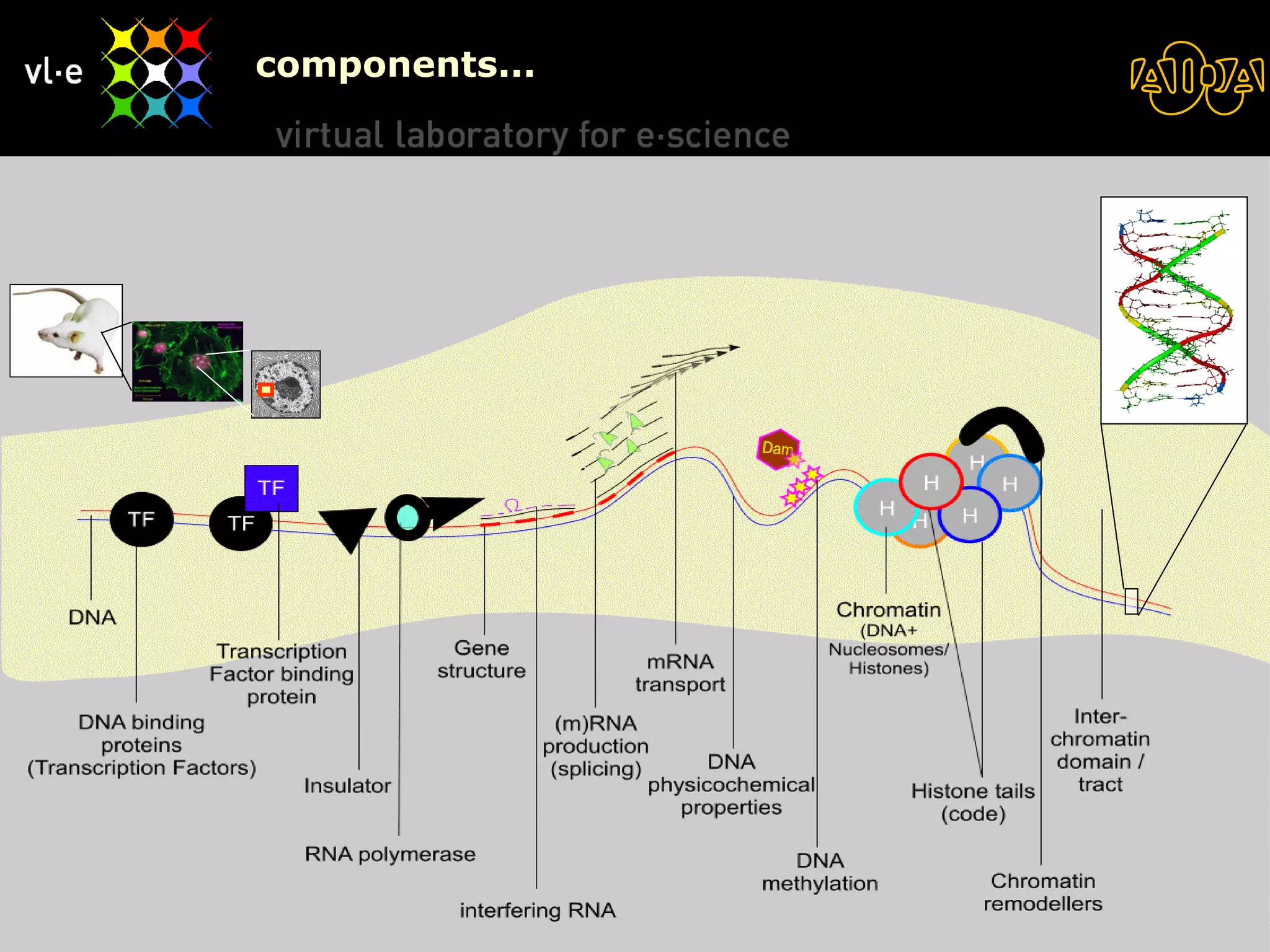
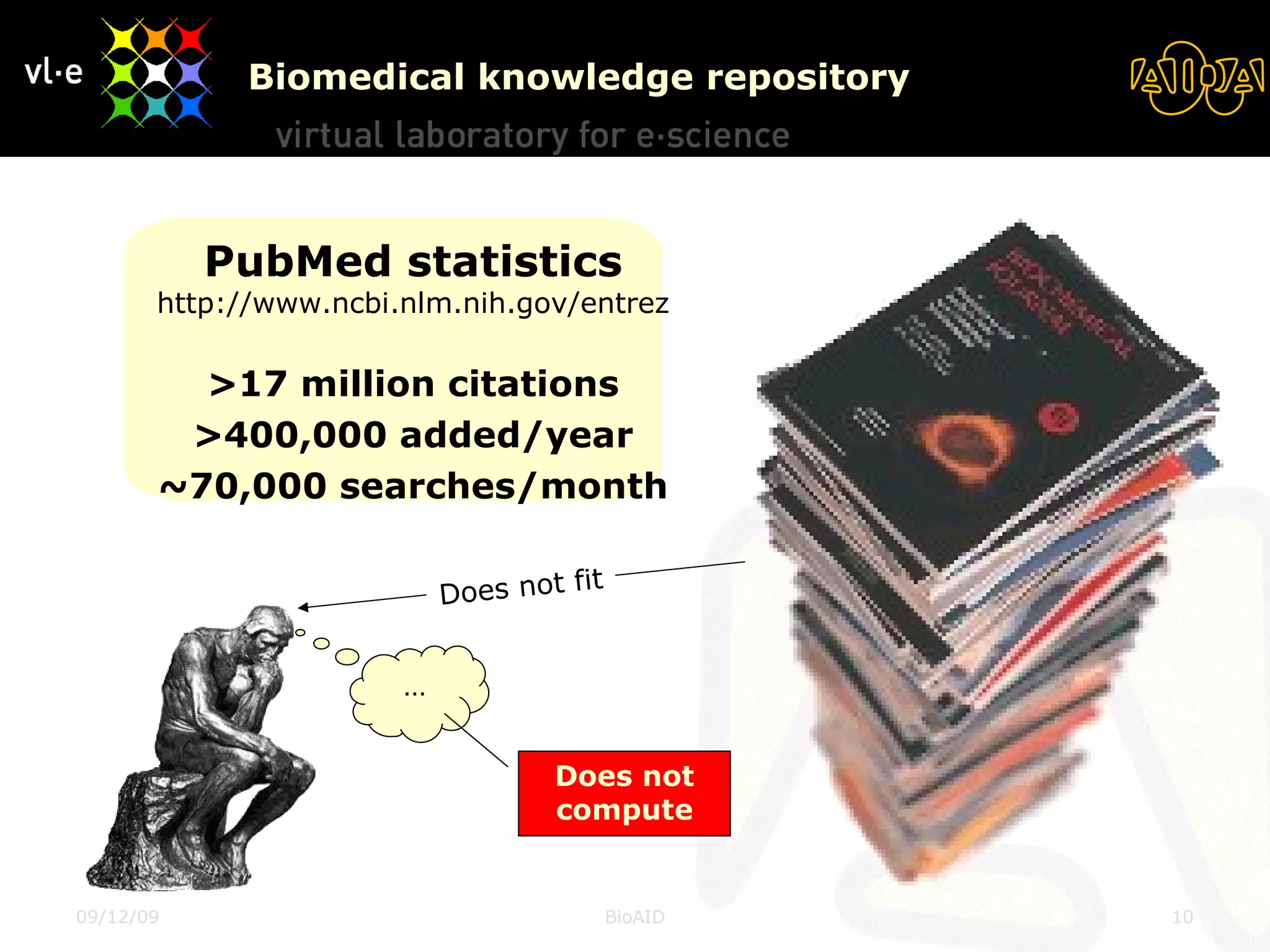
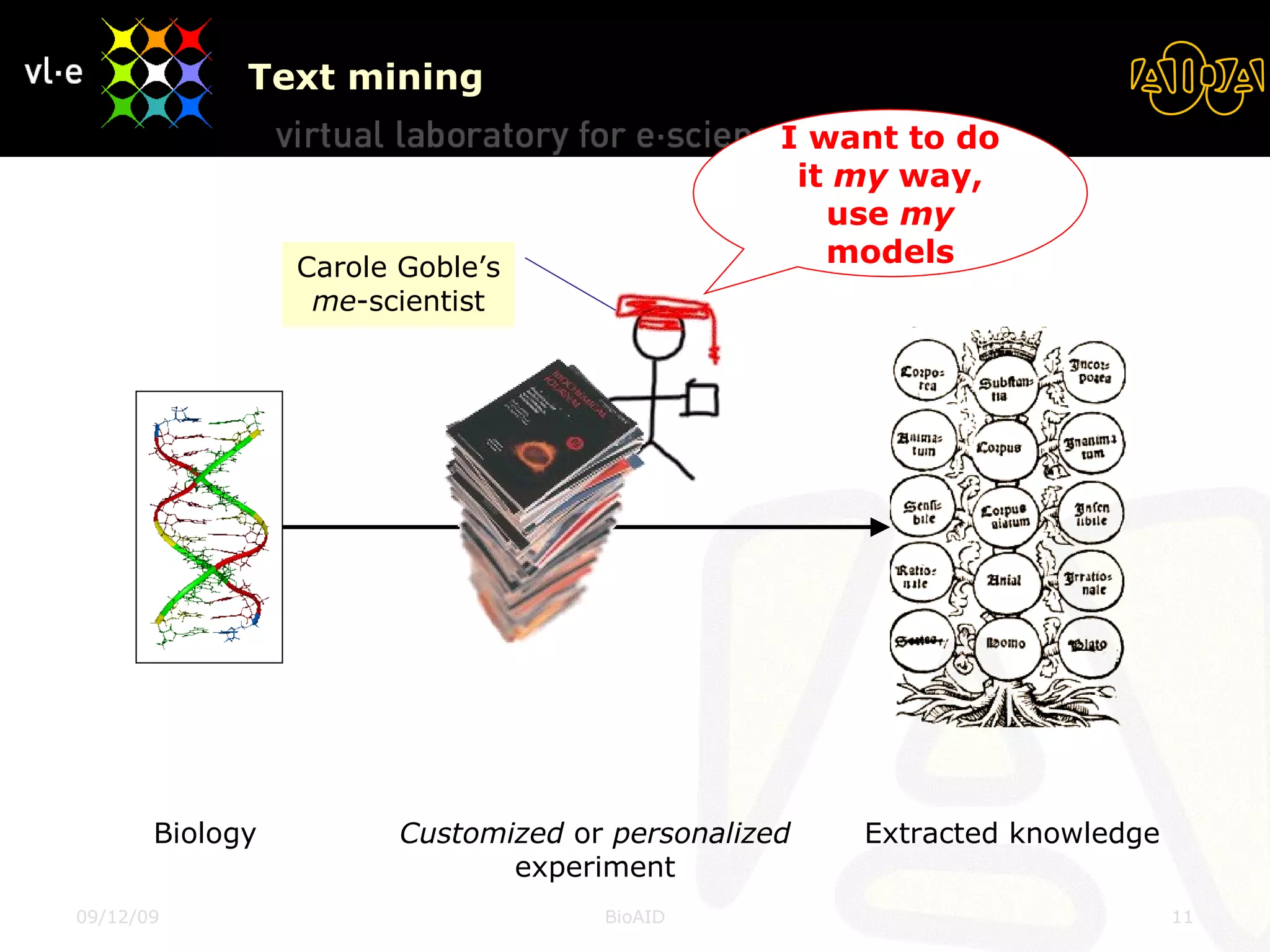
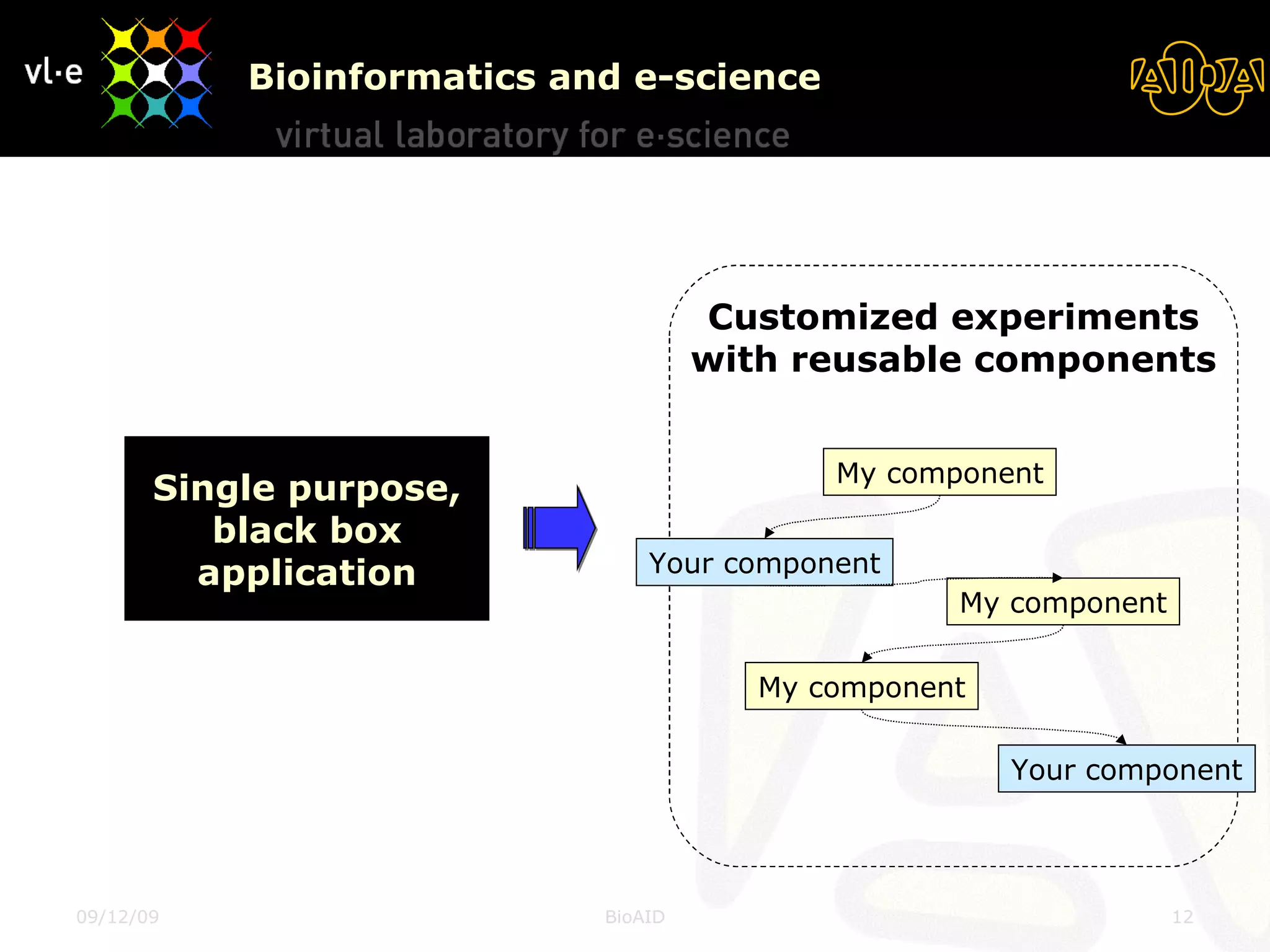
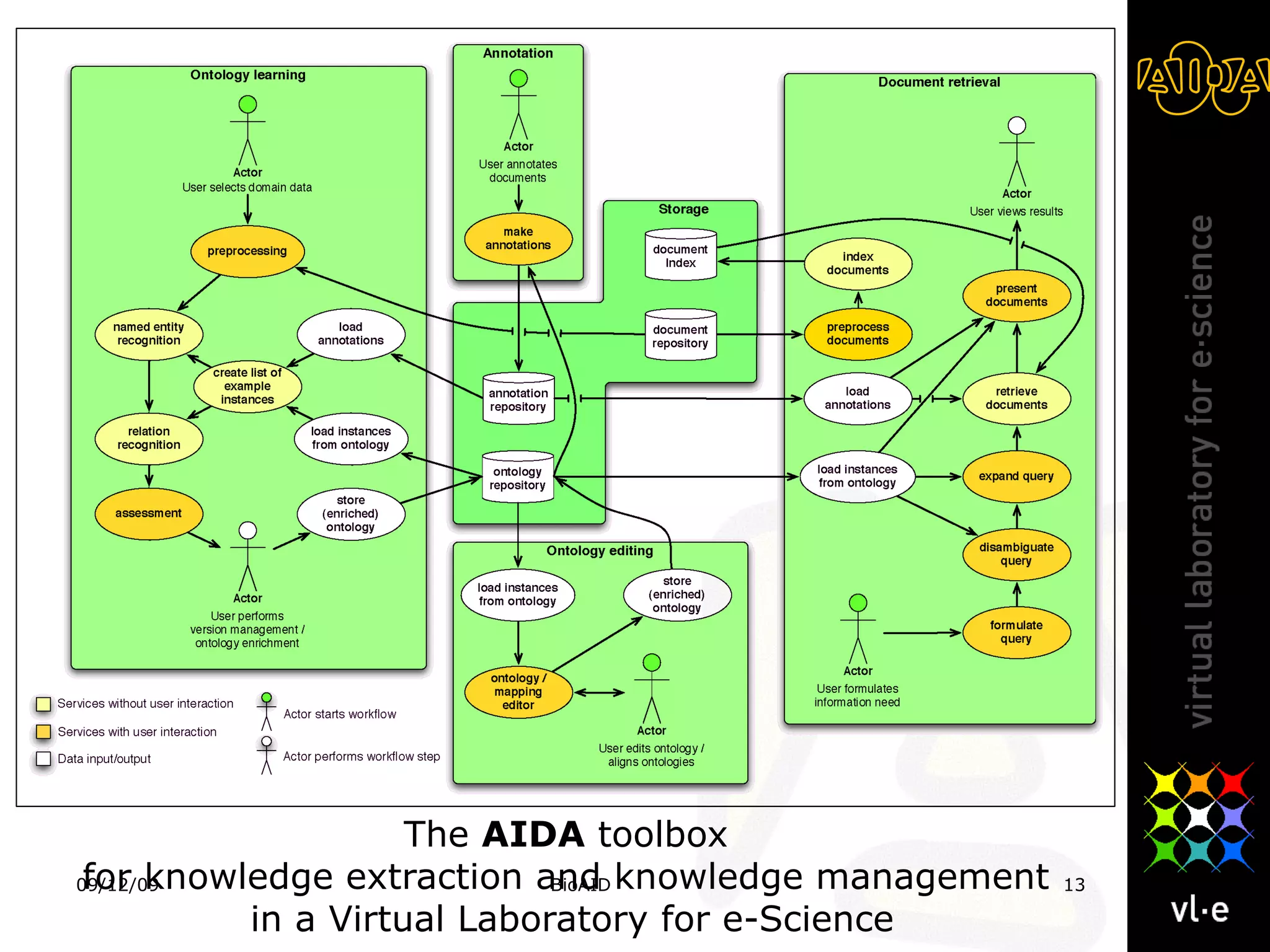
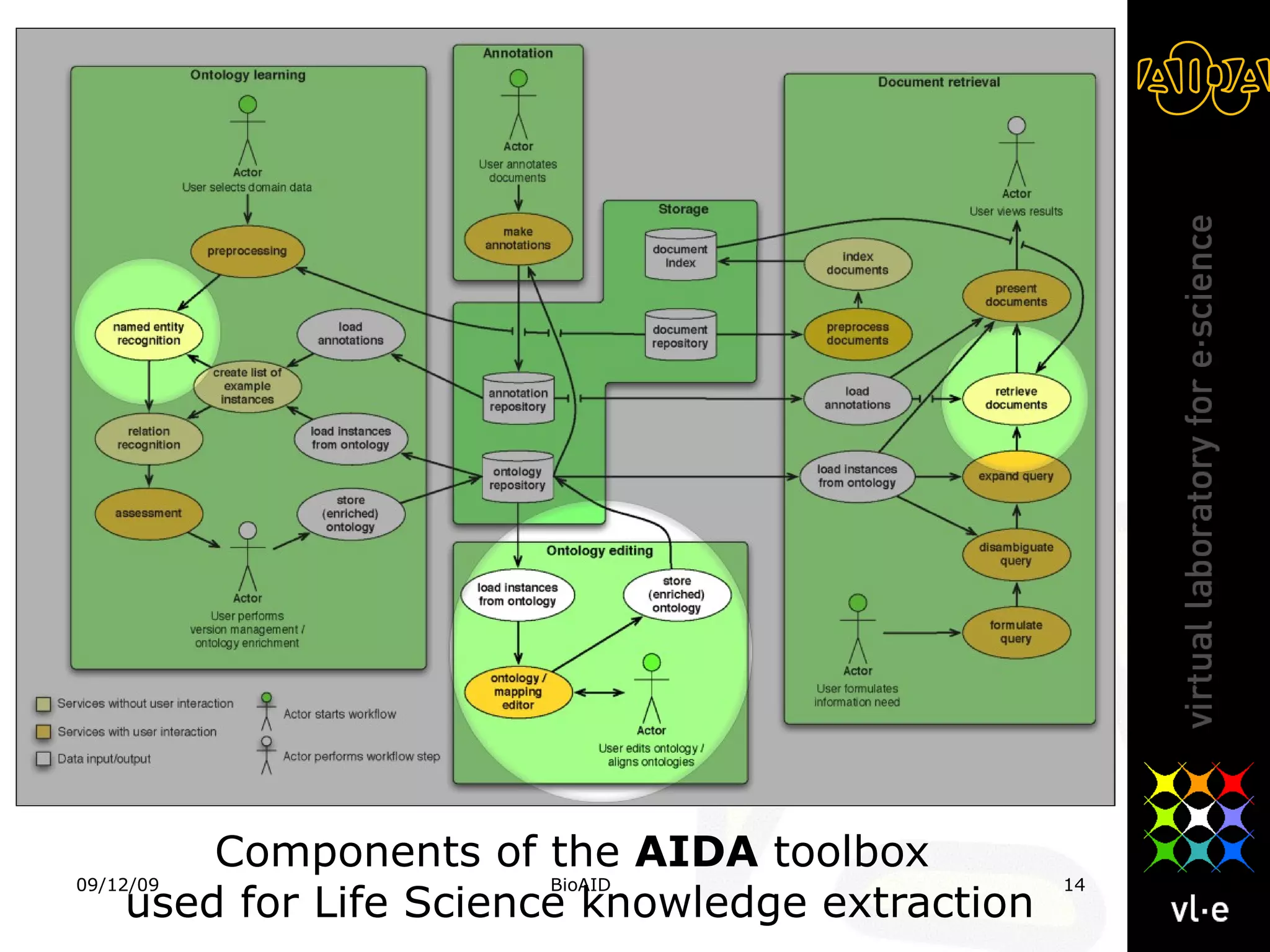
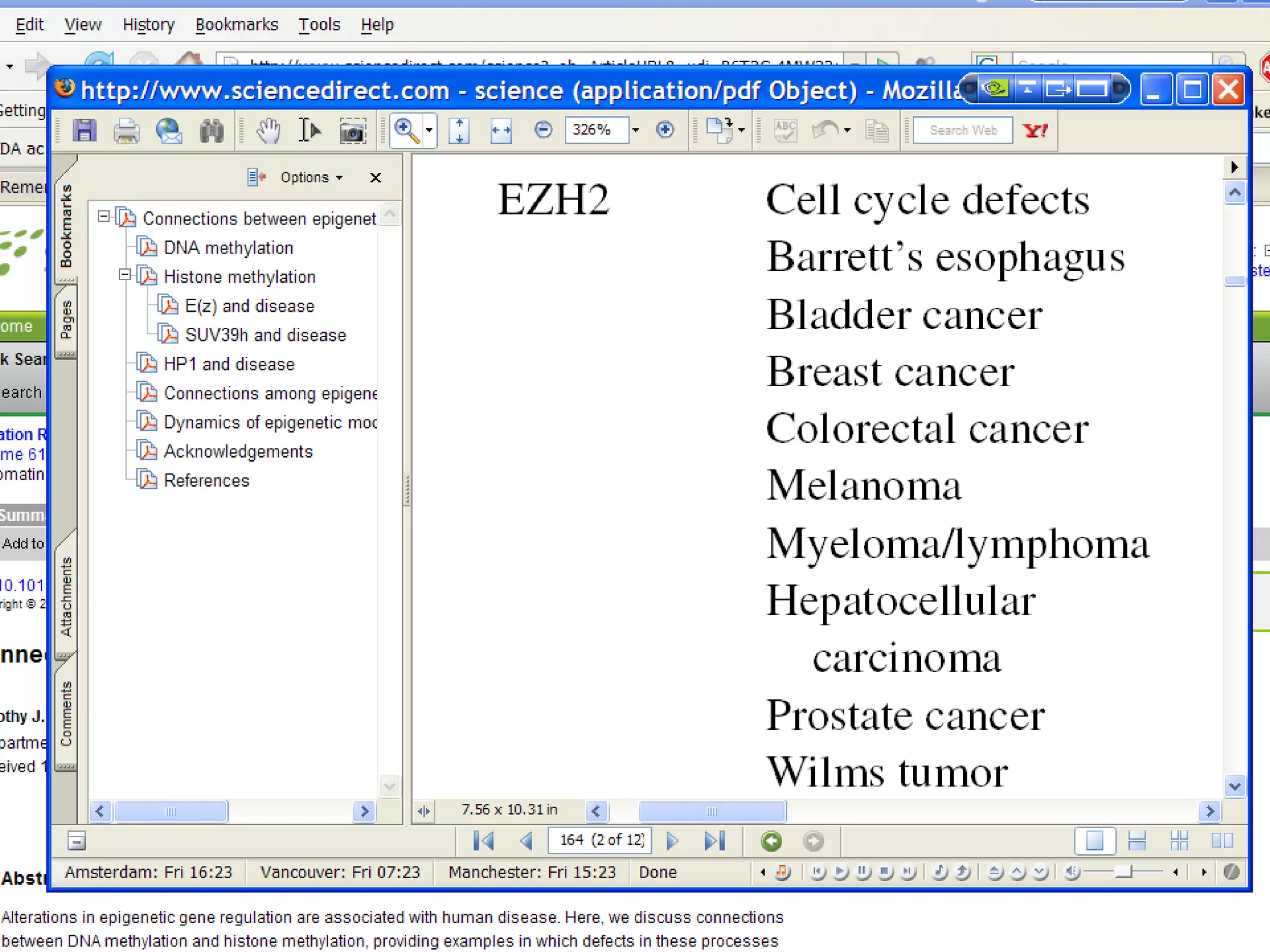
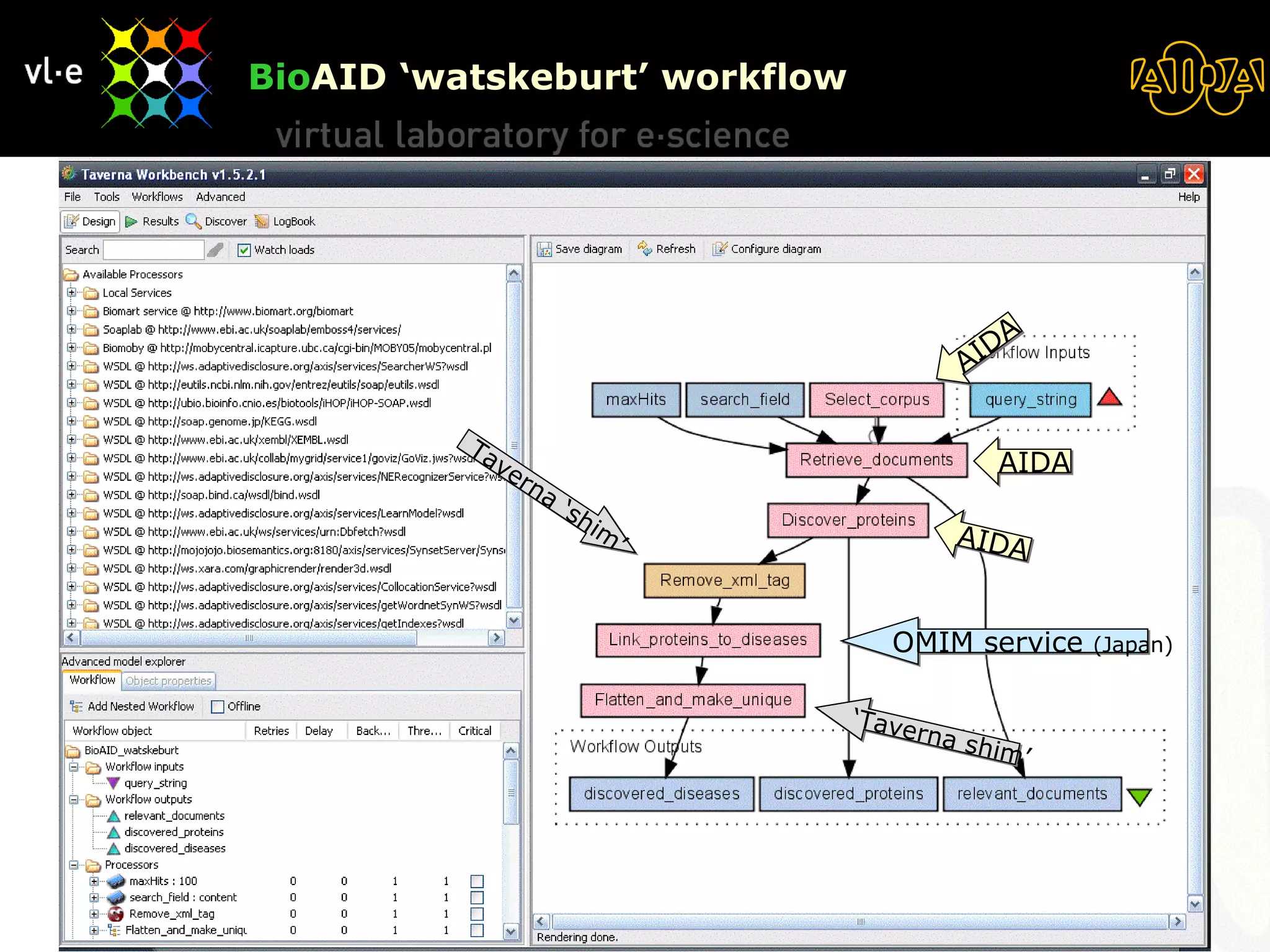
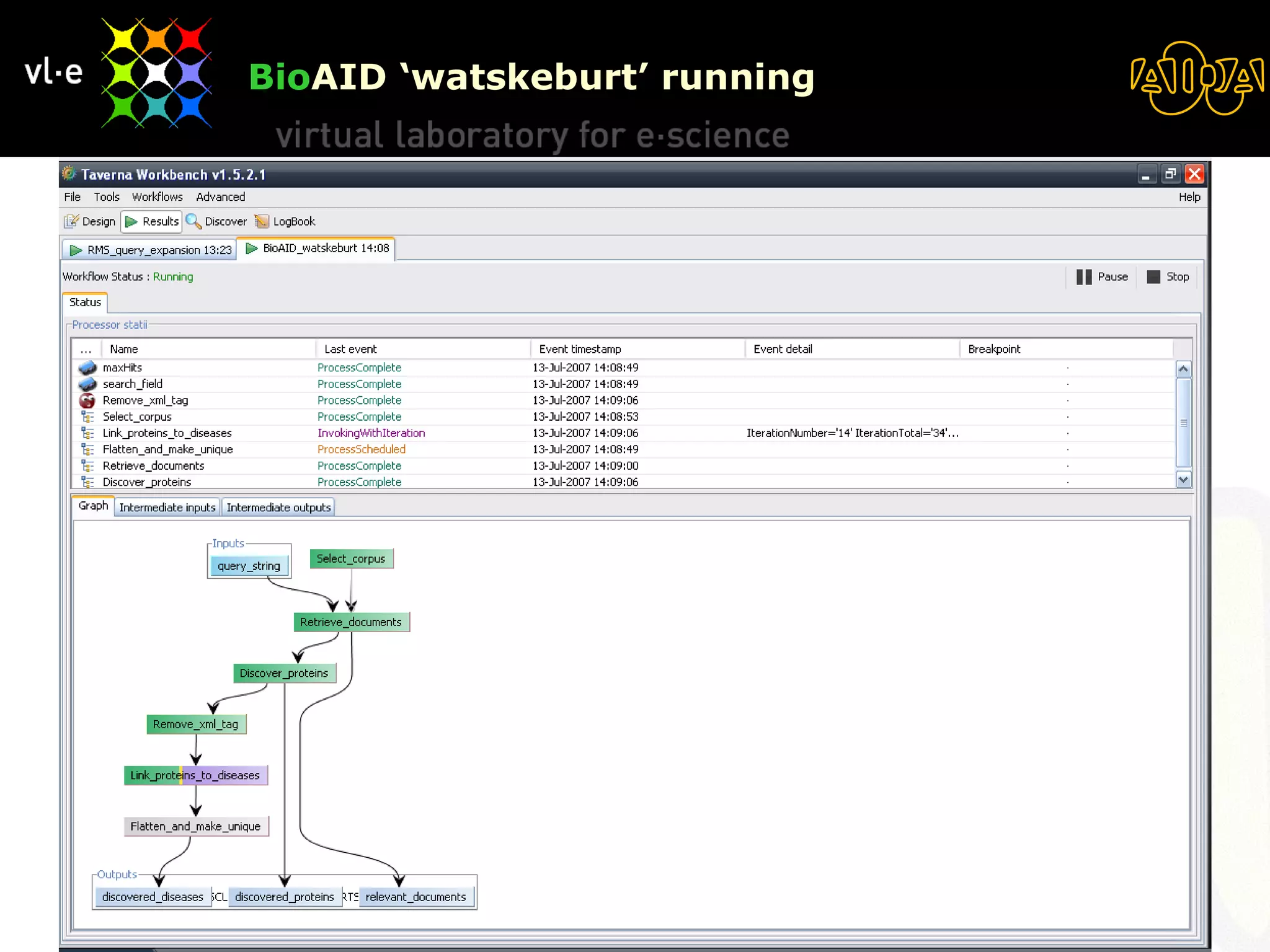
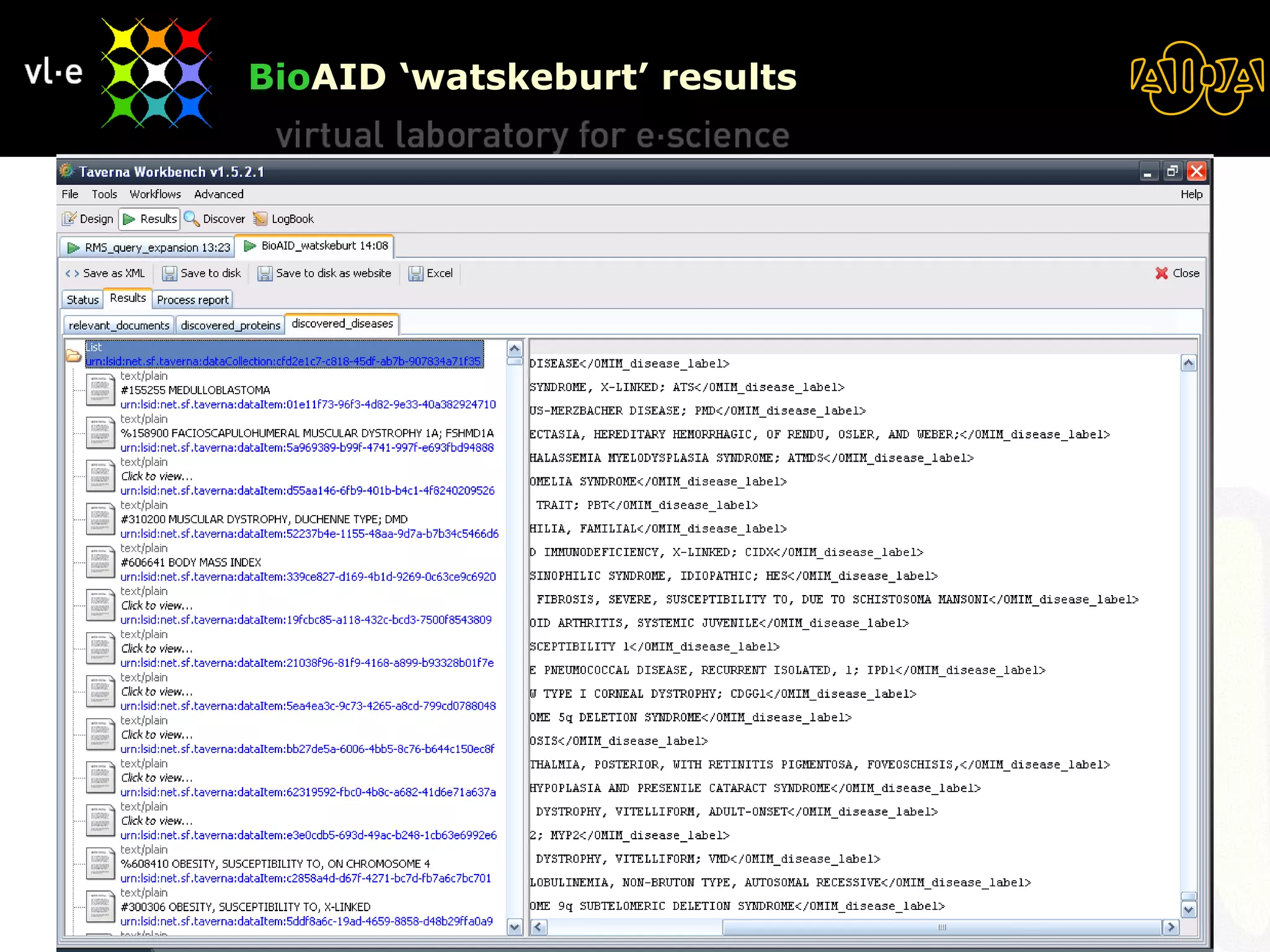
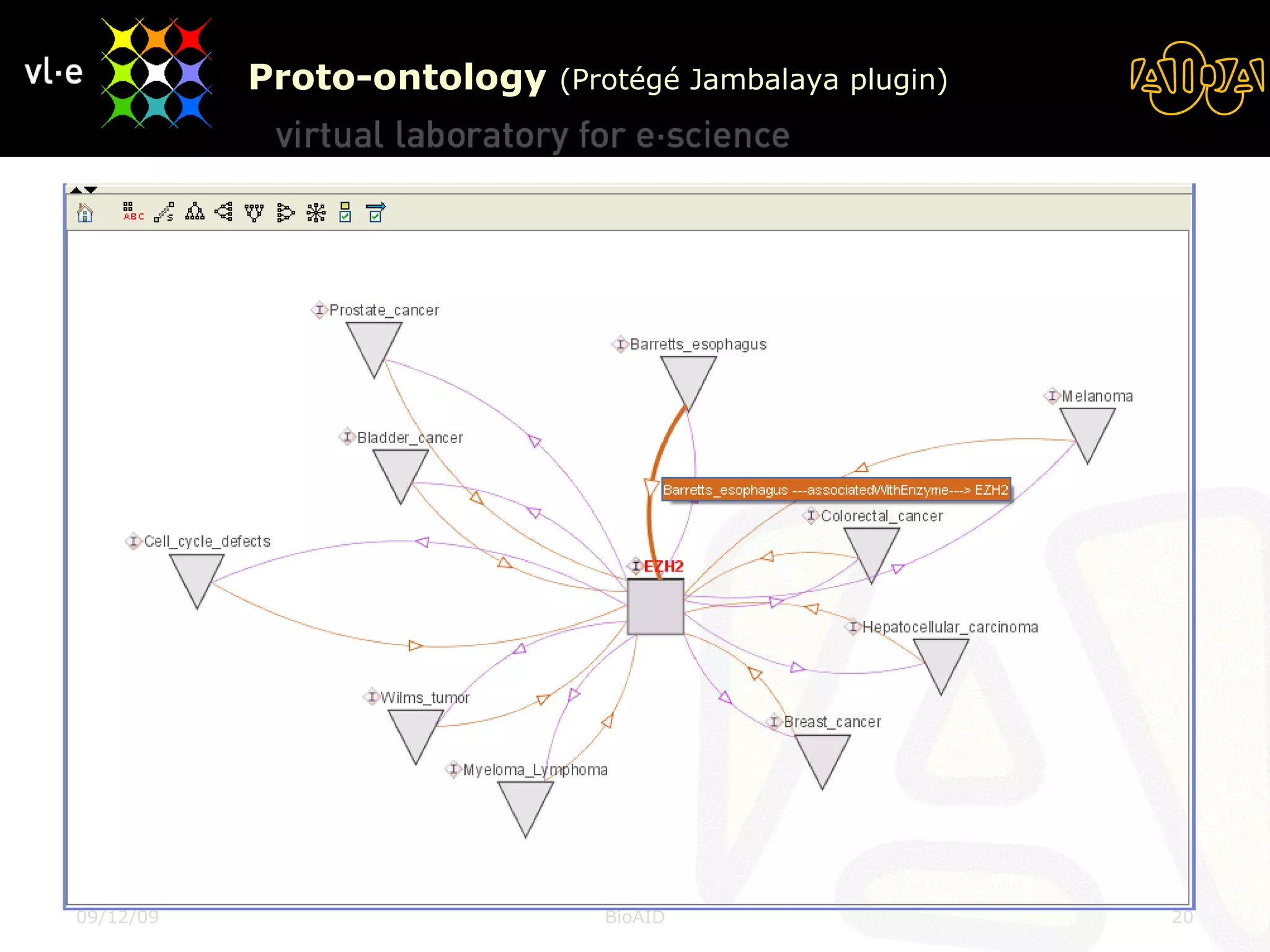
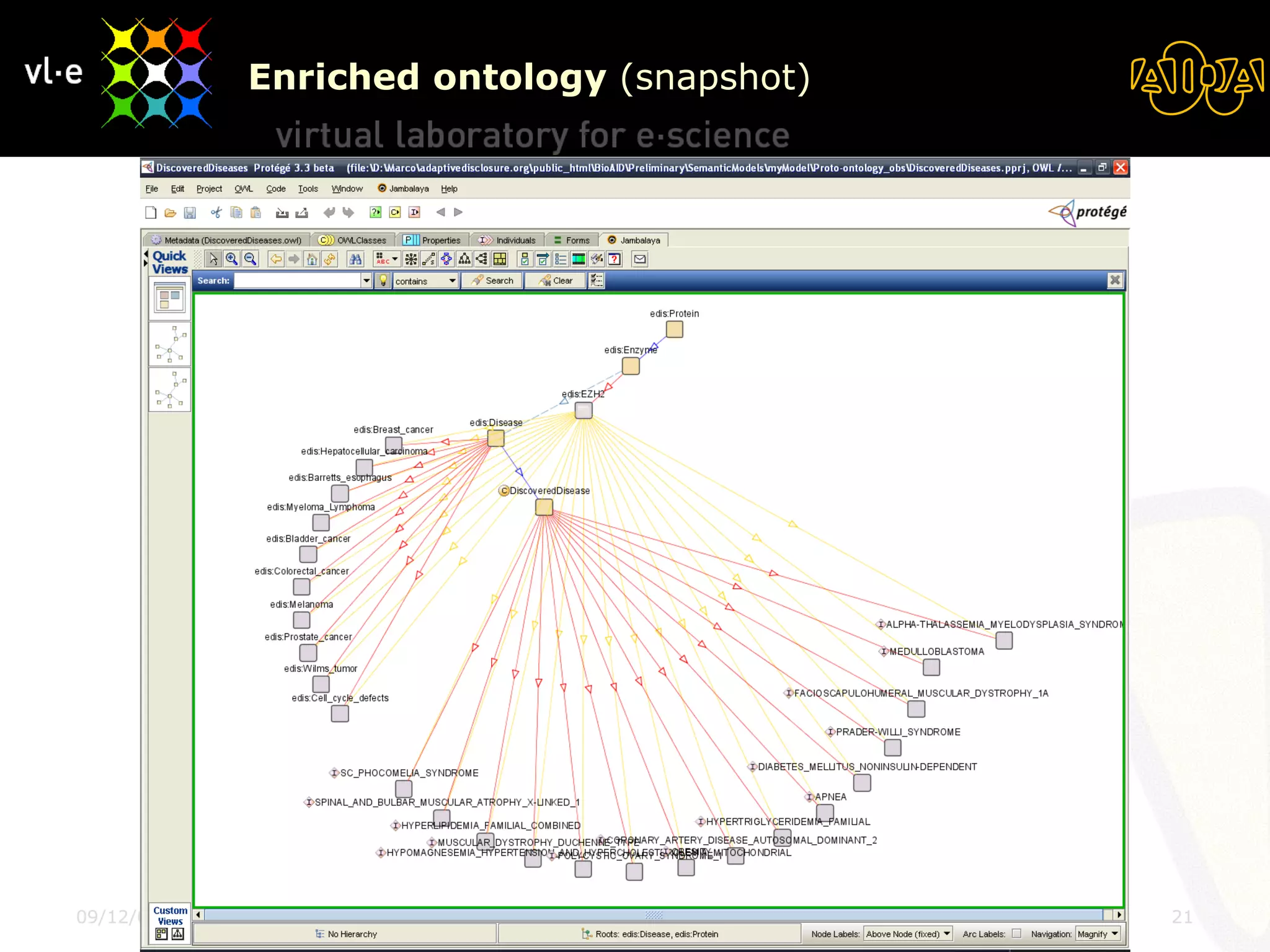
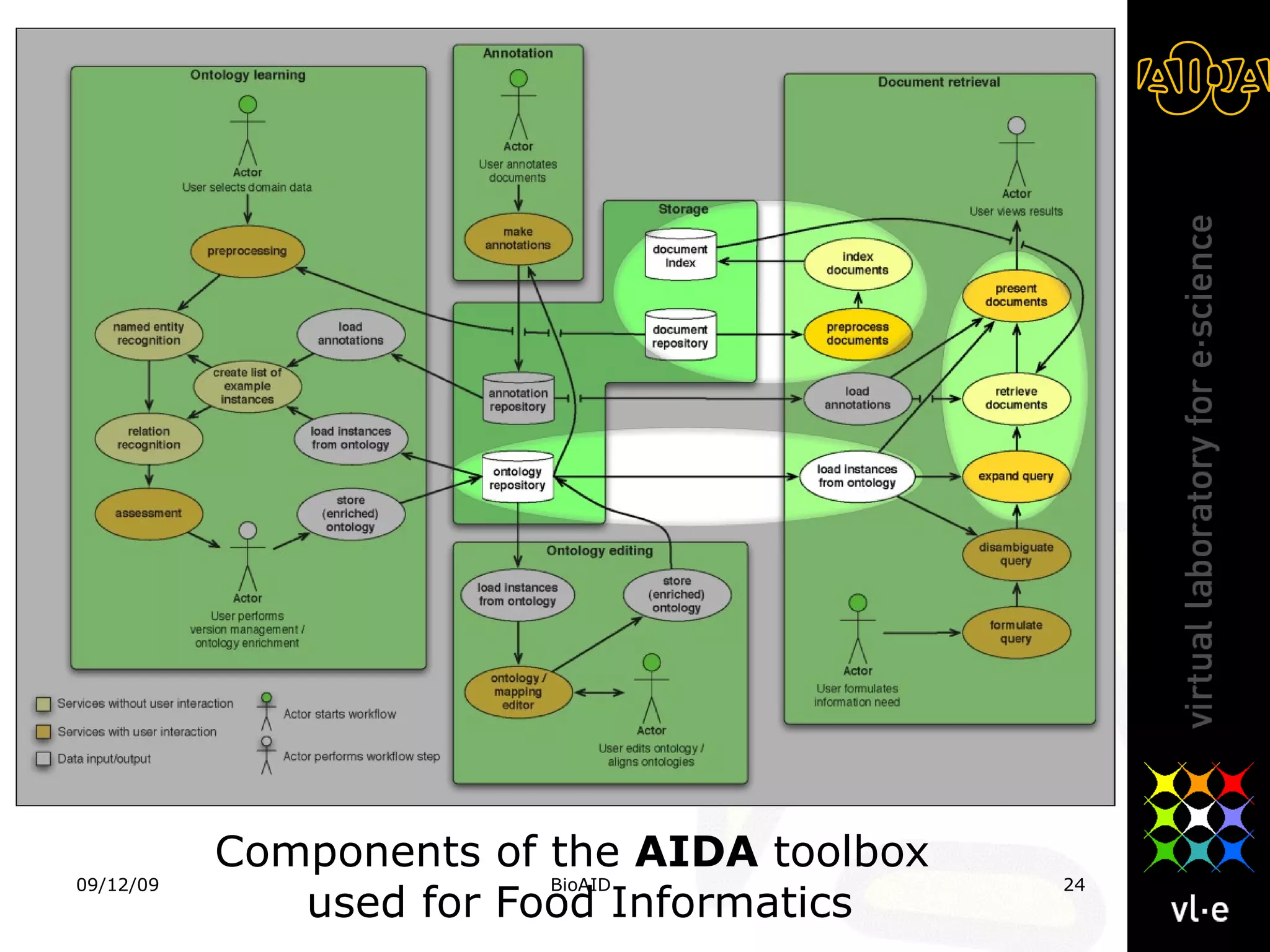
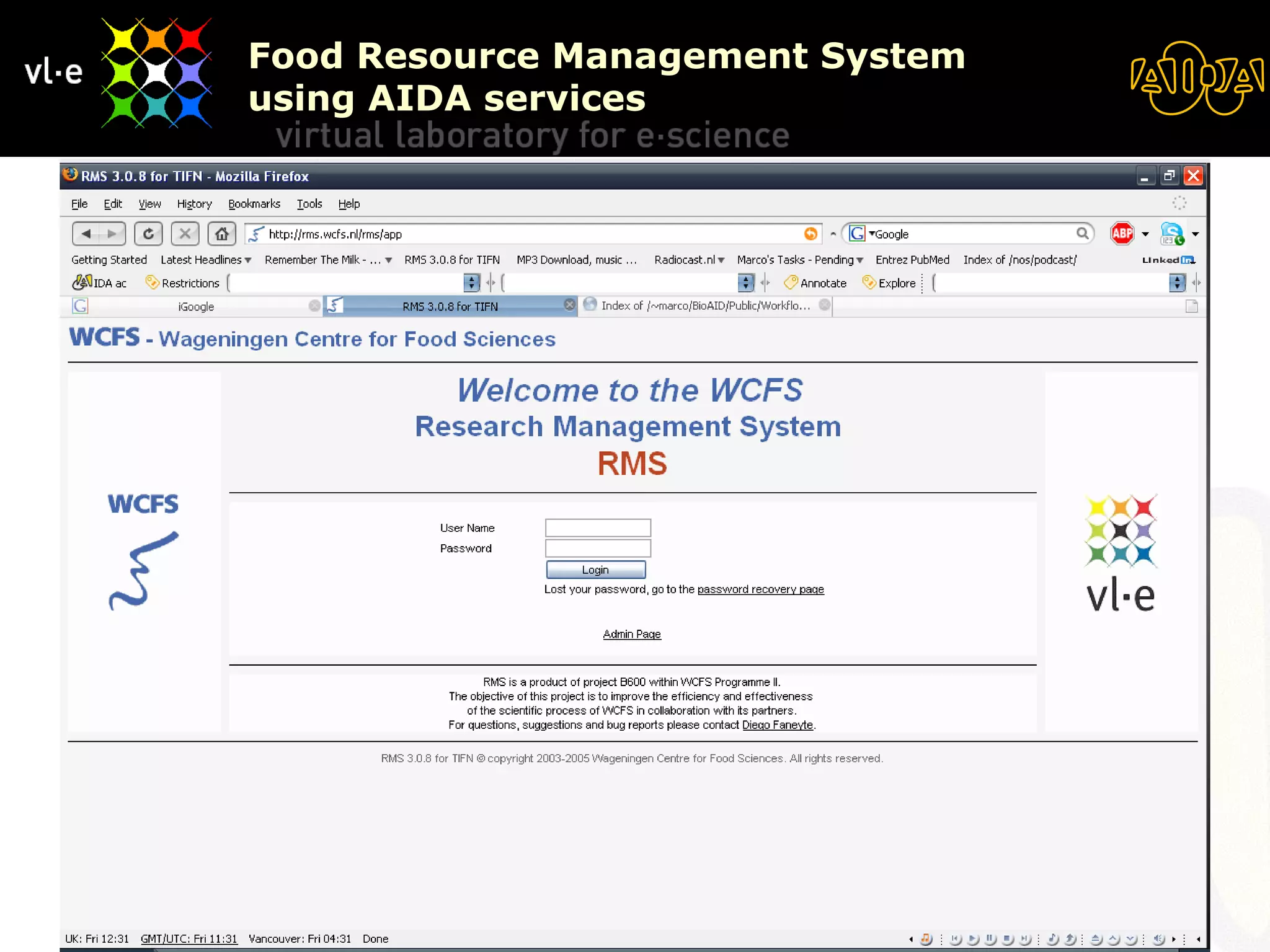
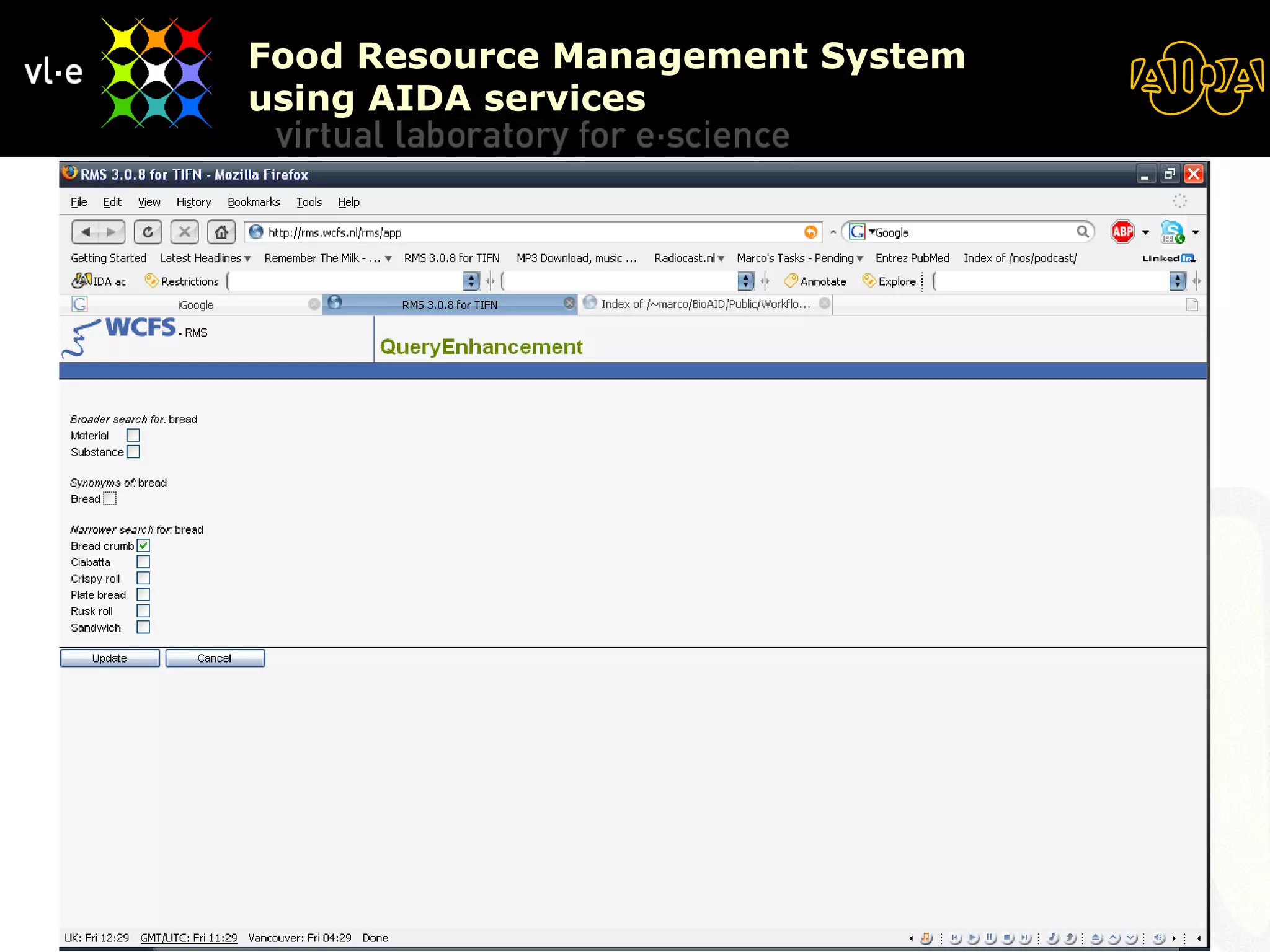
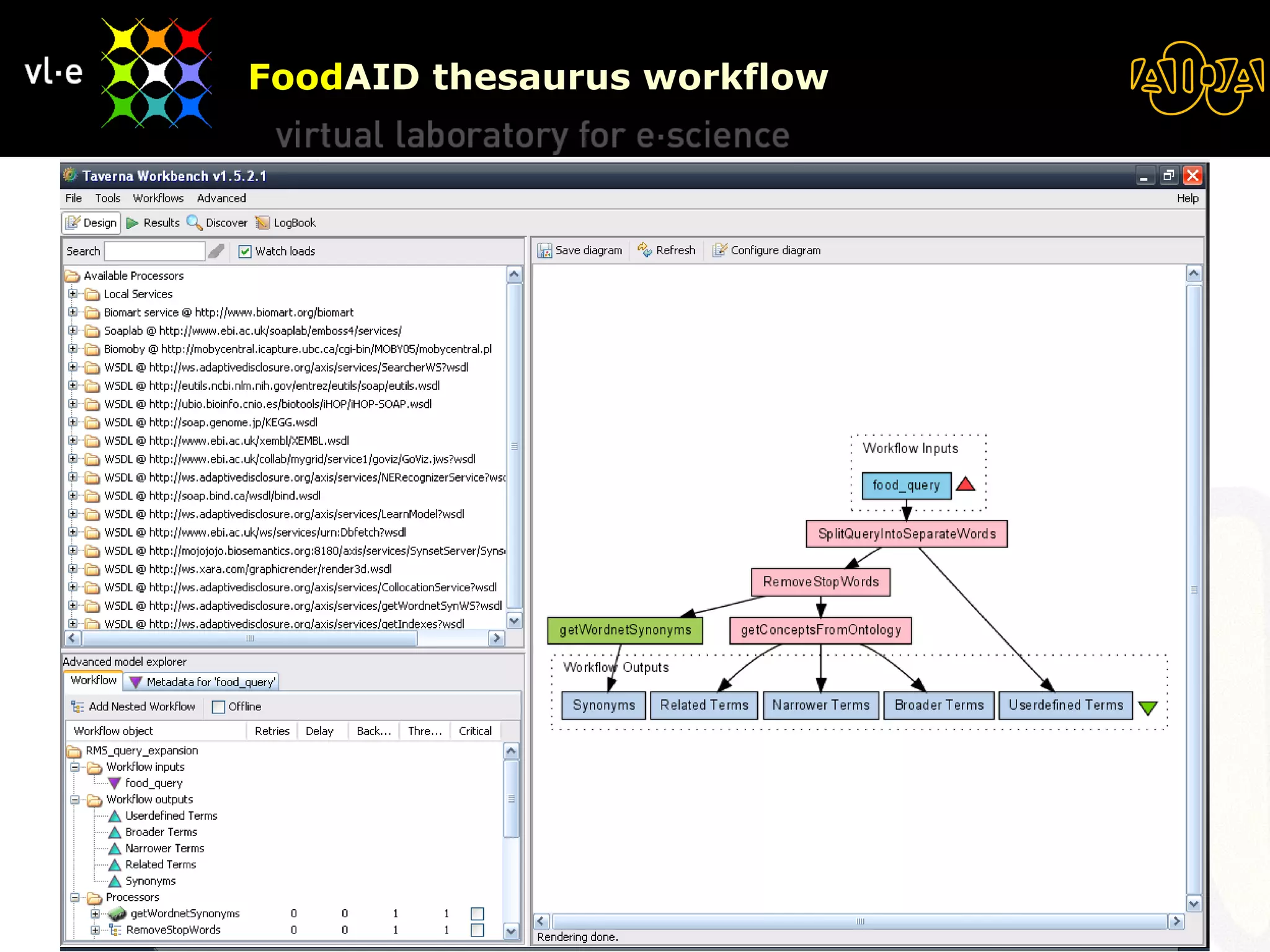
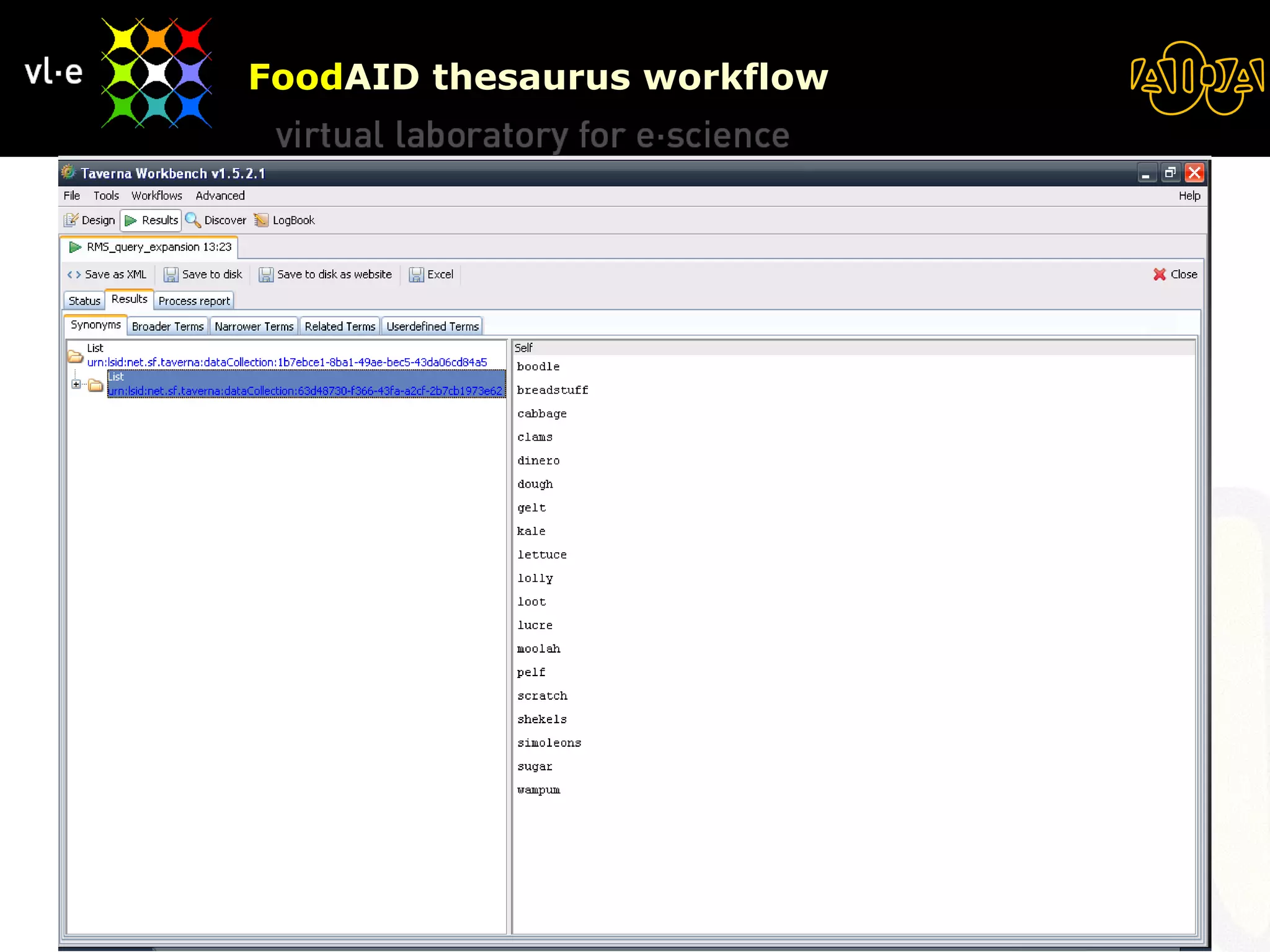
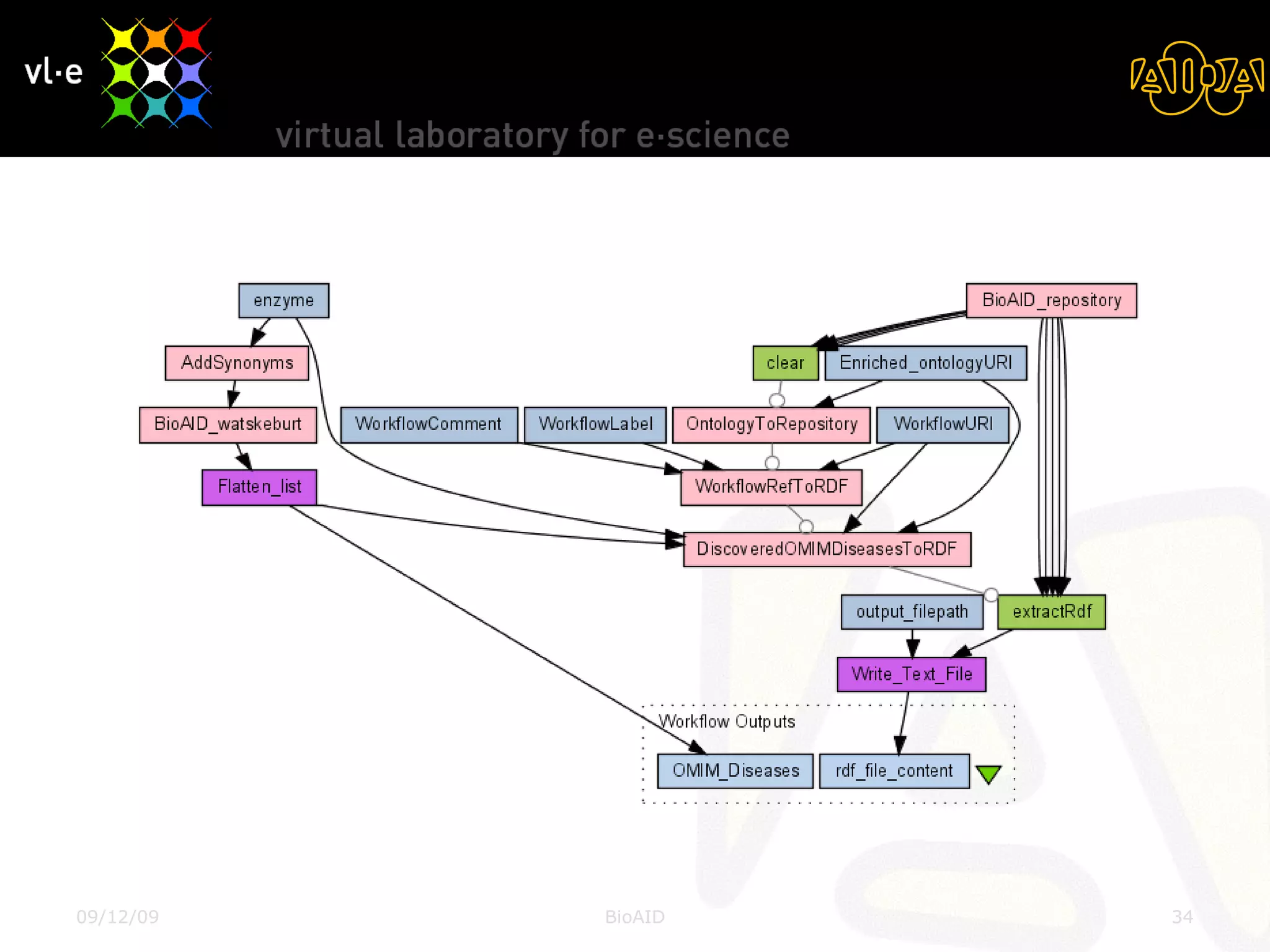
The document summarizes a presentation about AIDA, a biological knowledge extraction toolbox used in a Virtual Laboratory for e-Science. The presentation demonstrated how AIDA can be used for life science applications like extracting knowledge from literature on chromatin structure, and for food informatics applications like developing a food thesaurus. It also described components of the AIDA toolbox and examples of biological and food science workflows developed using AIDA services.