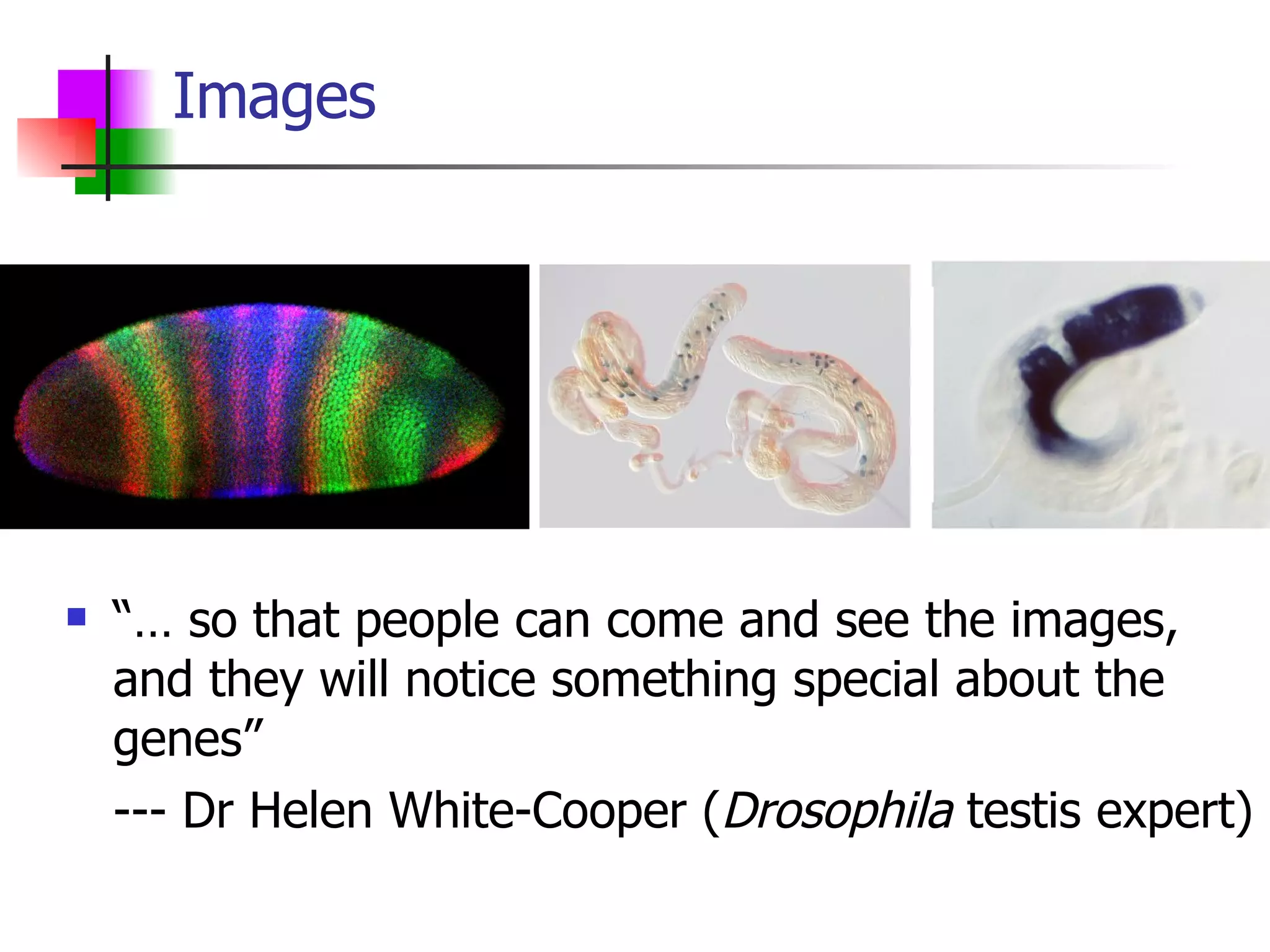
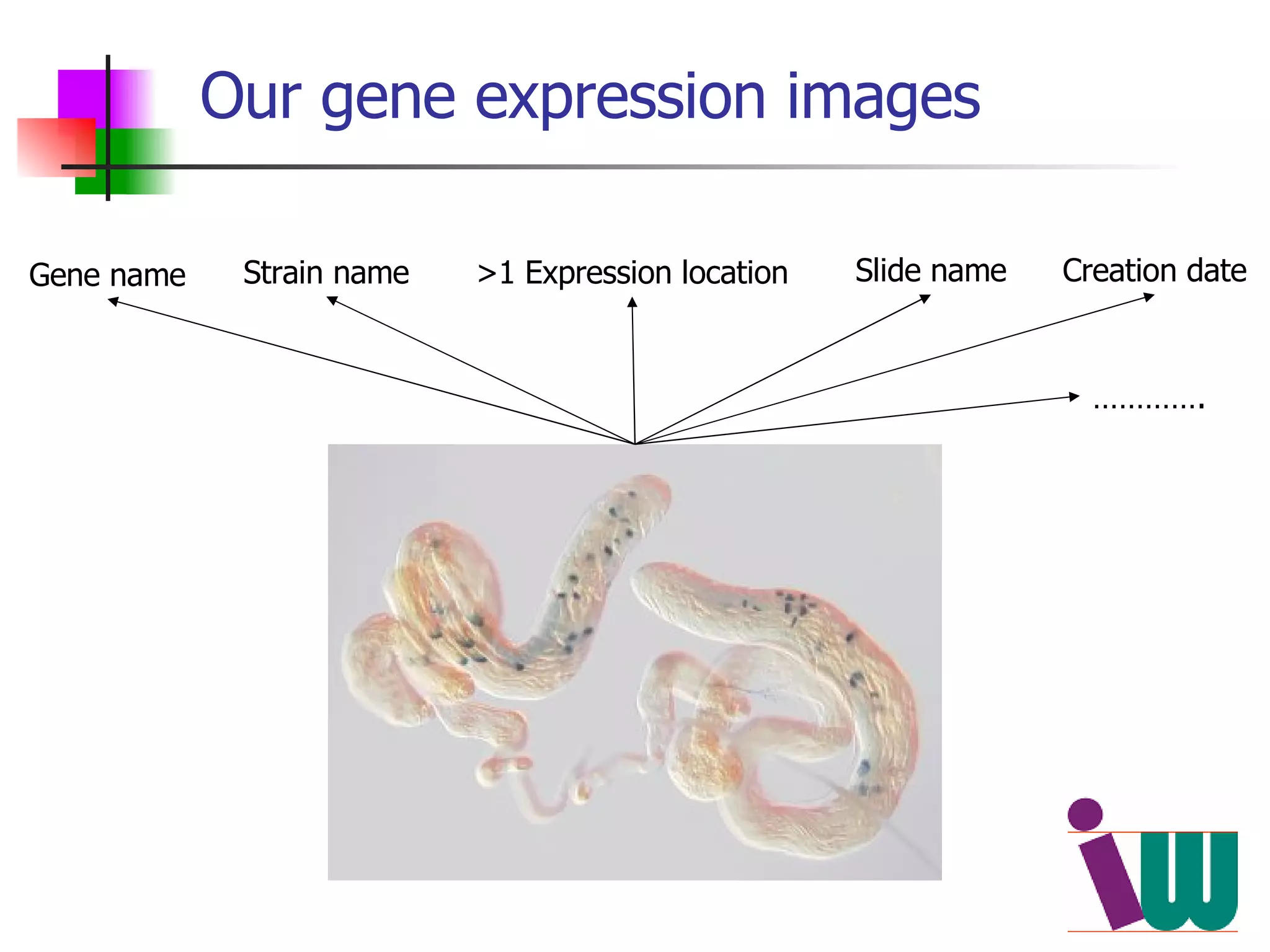
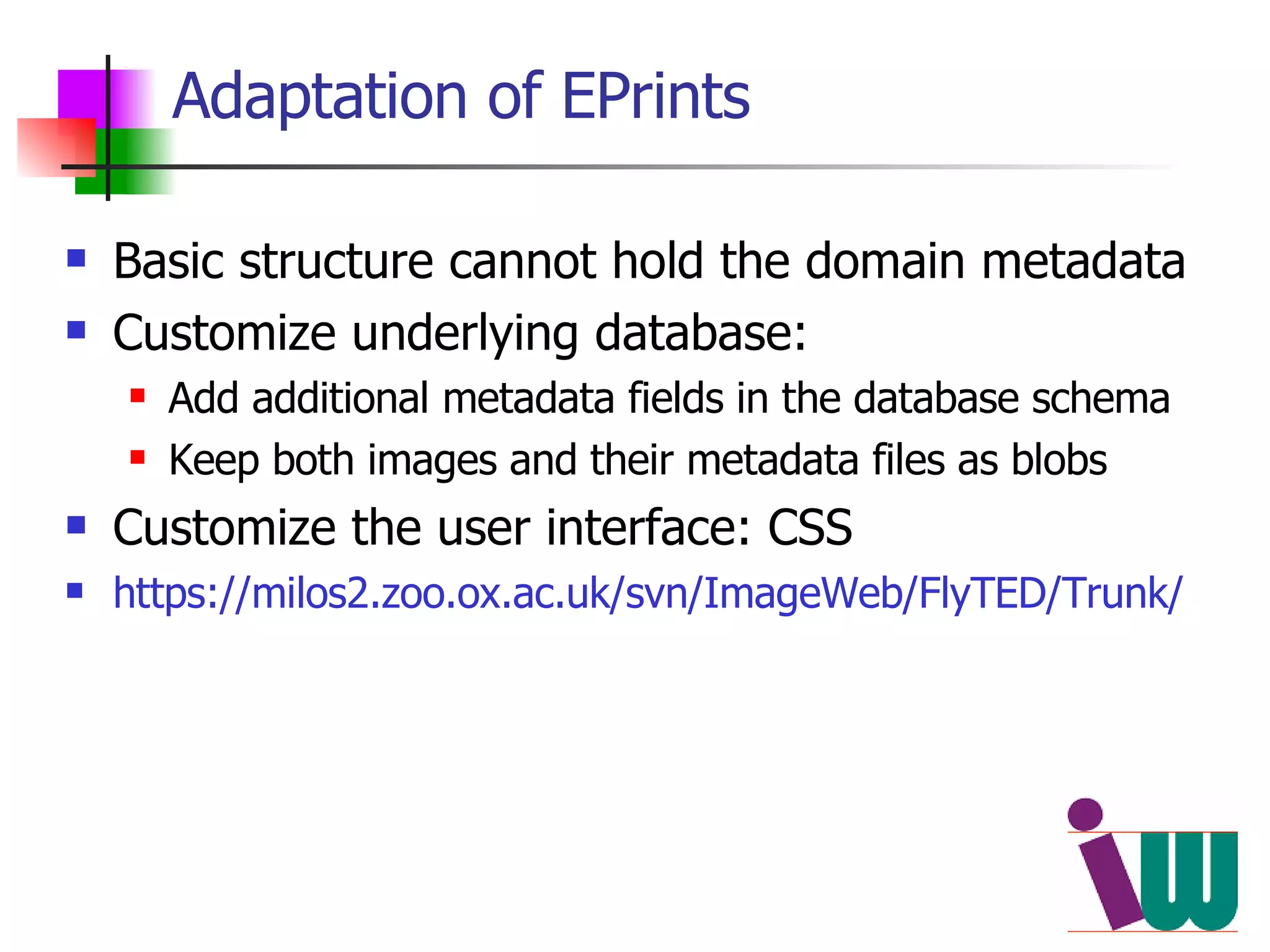
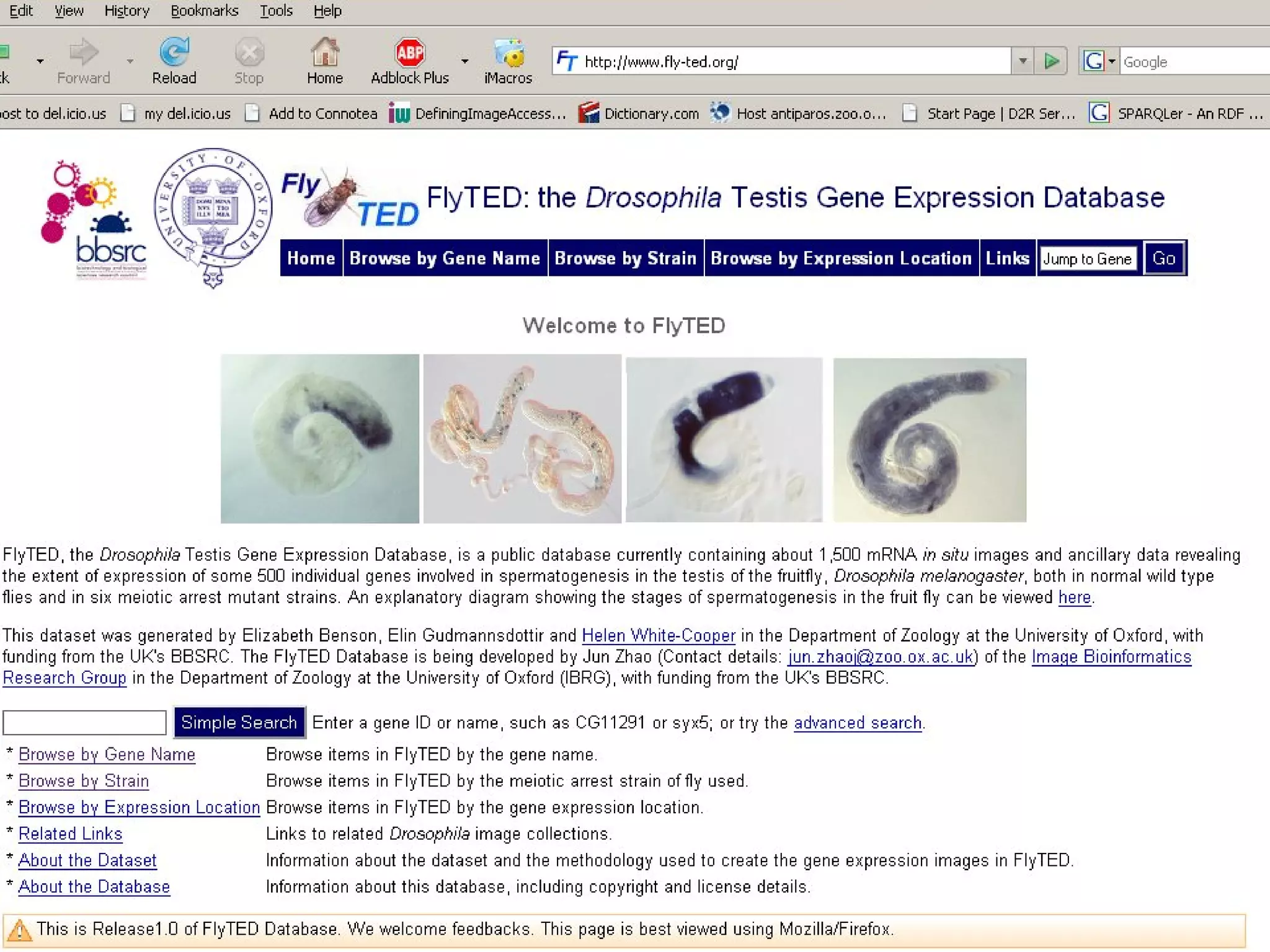
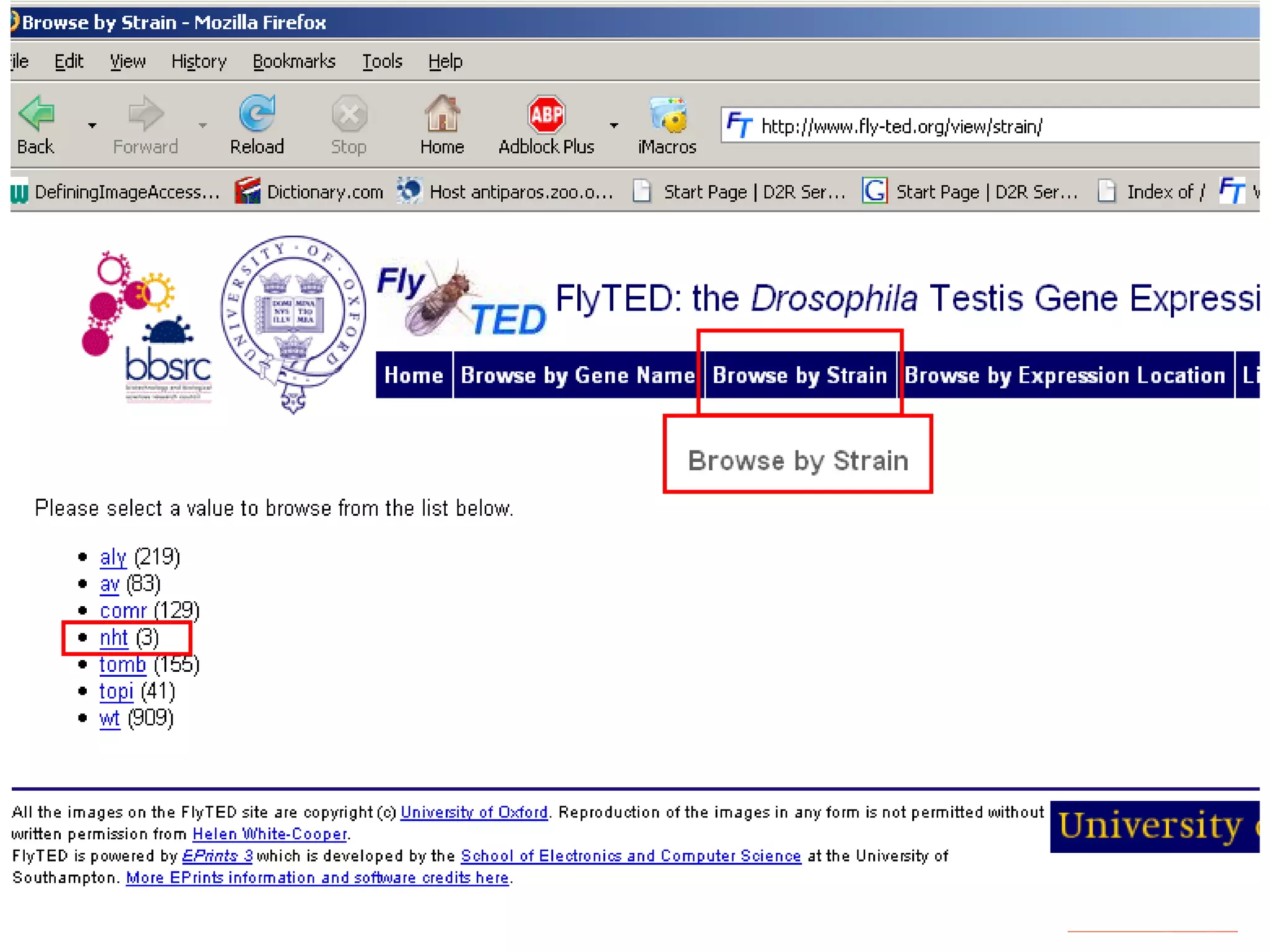
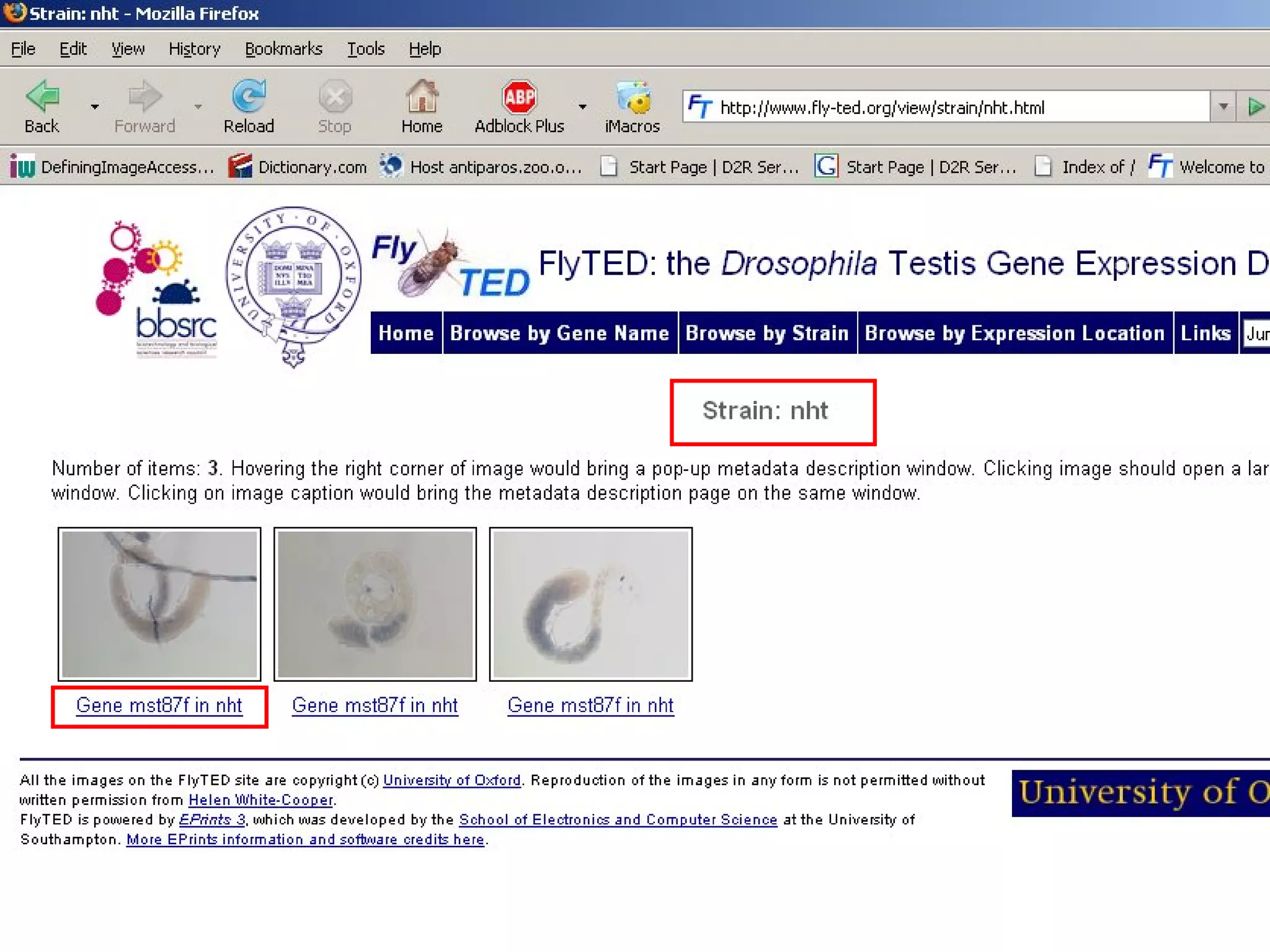
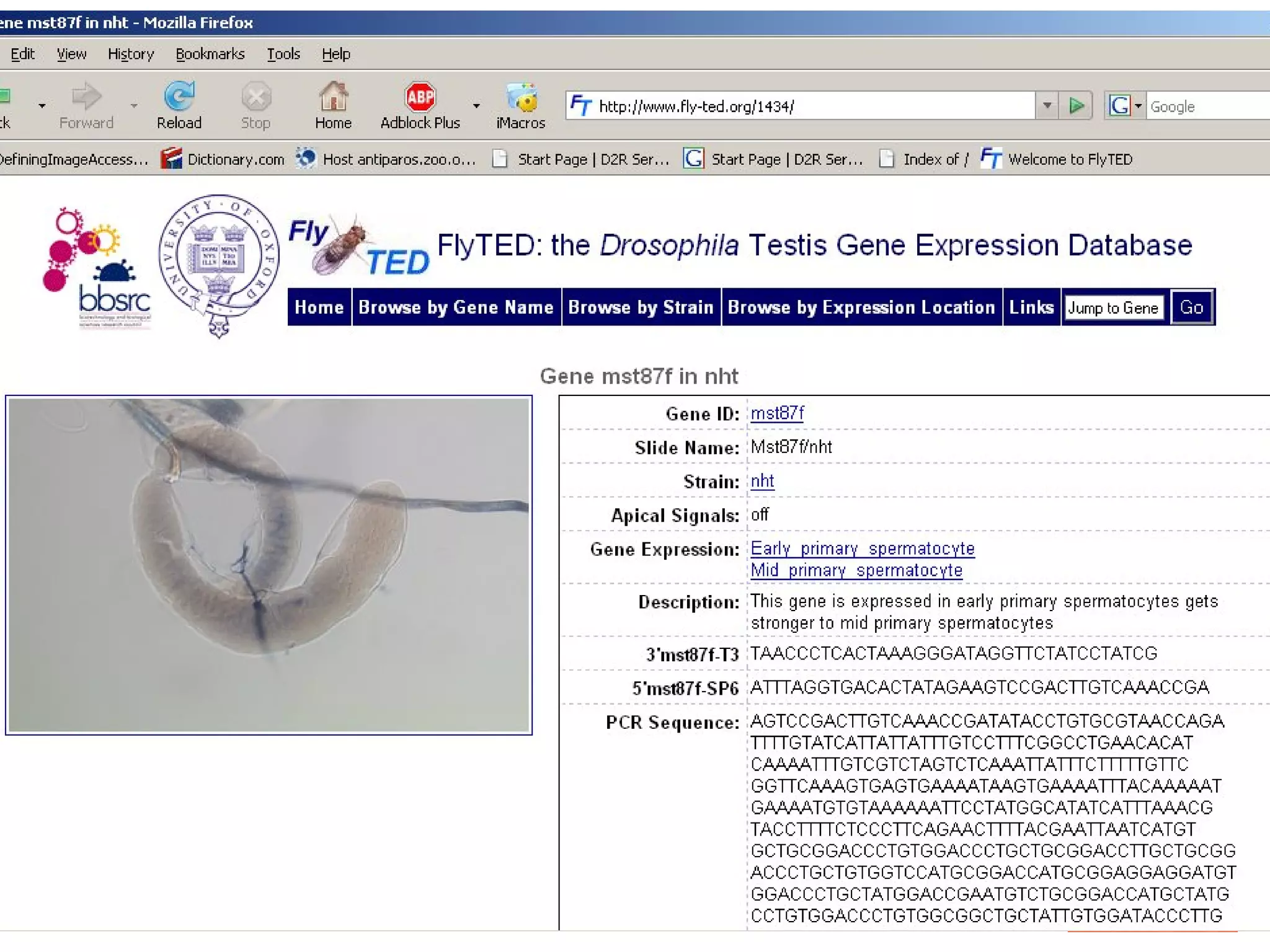
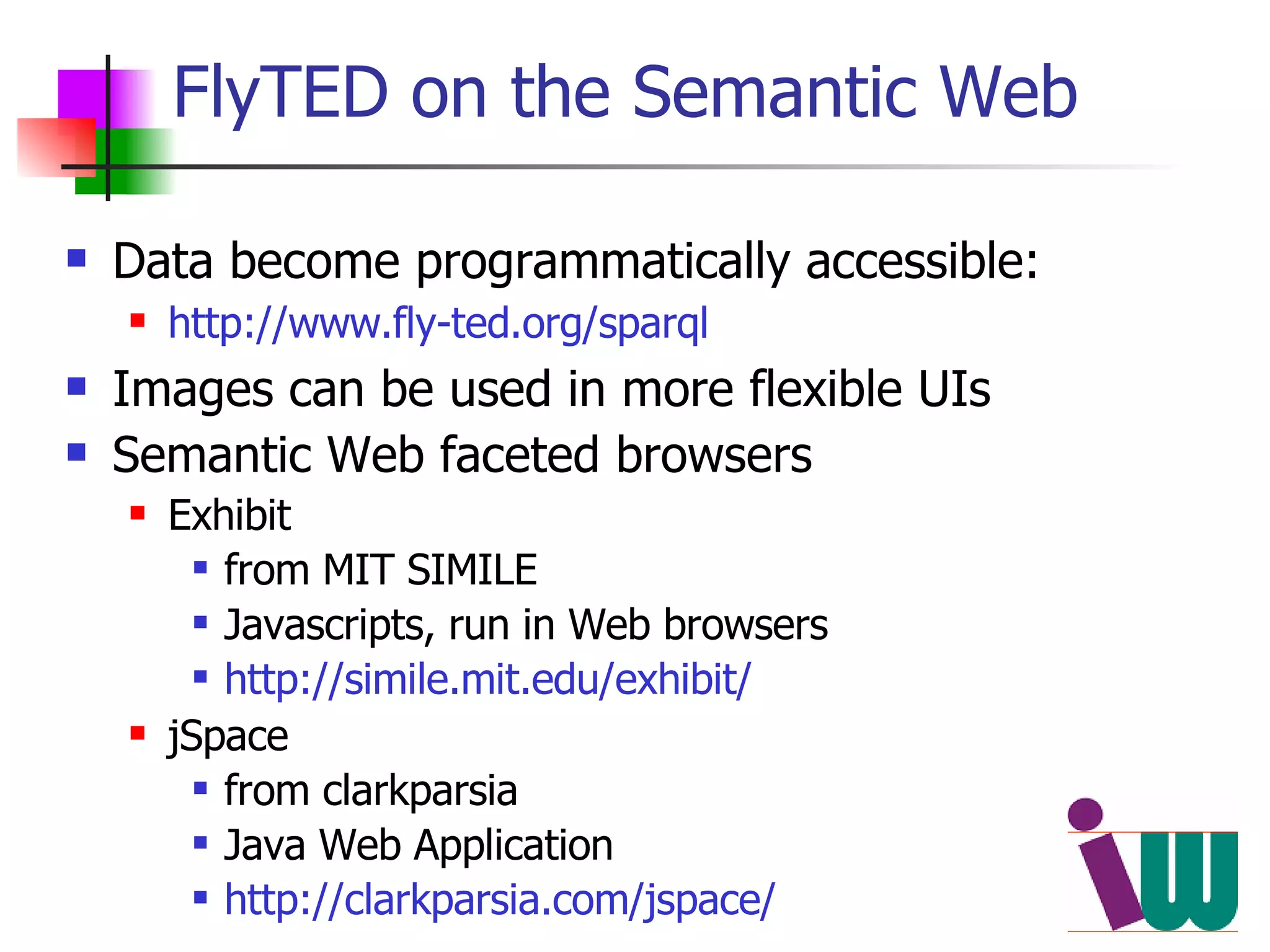
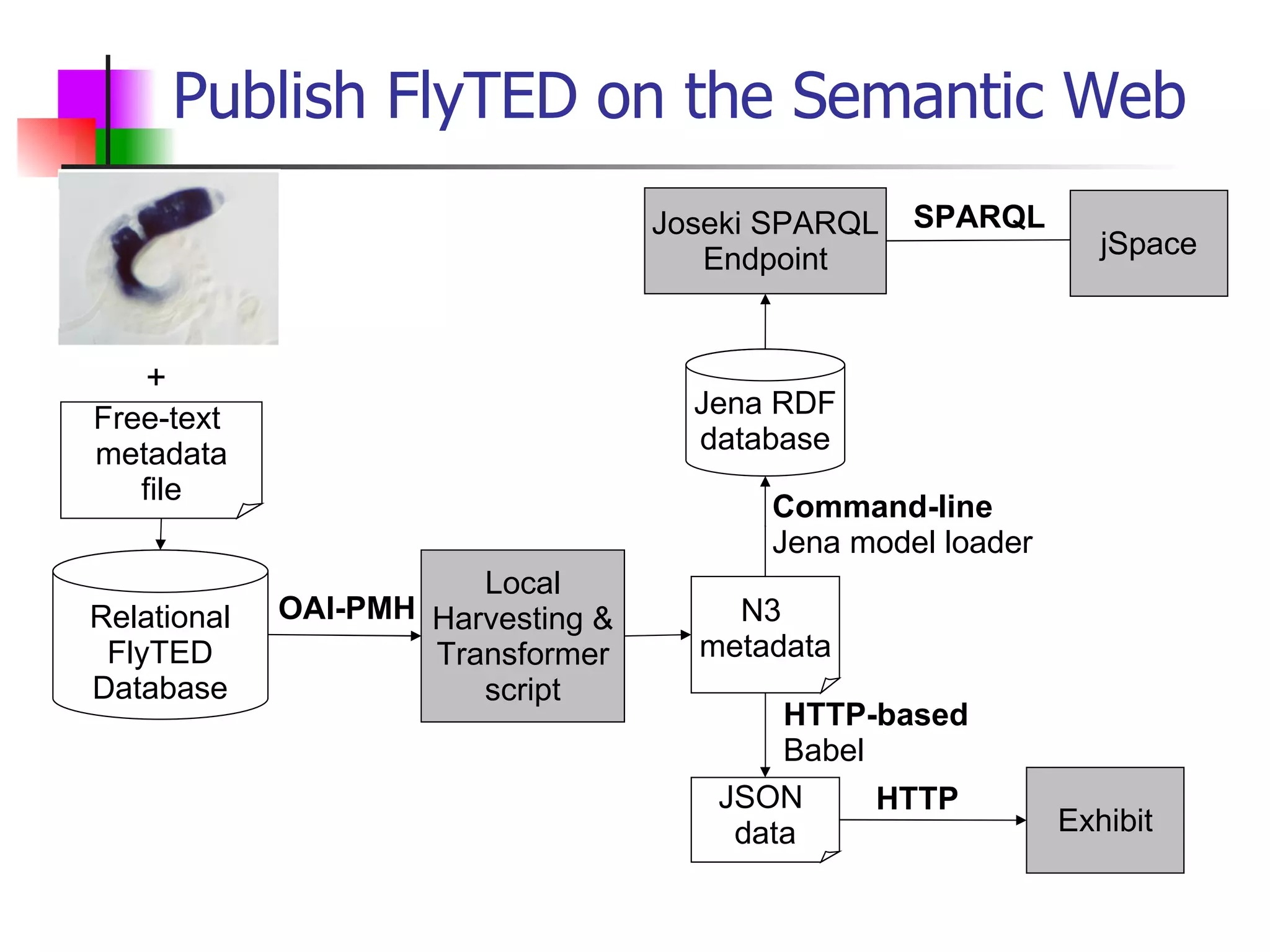
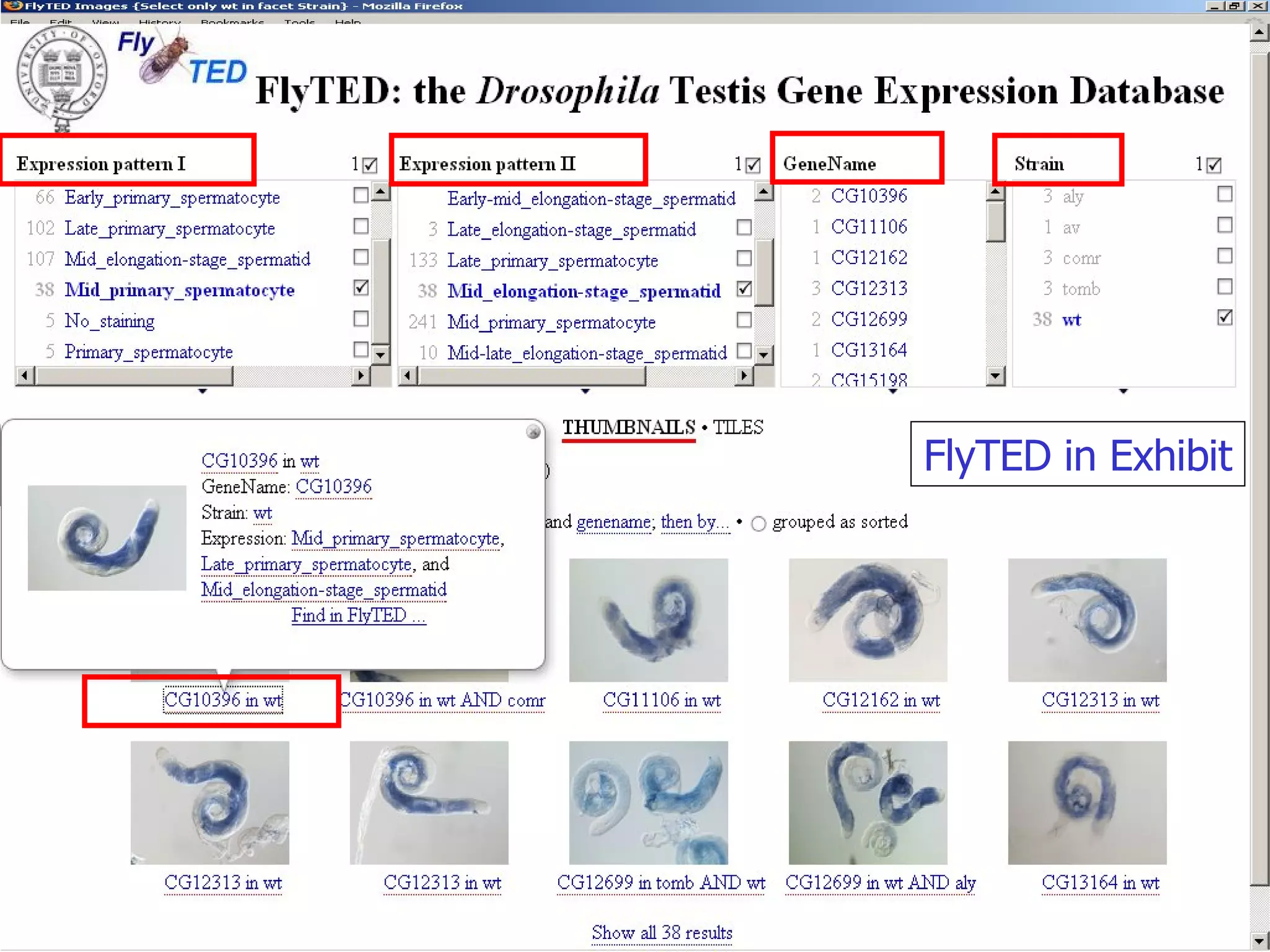
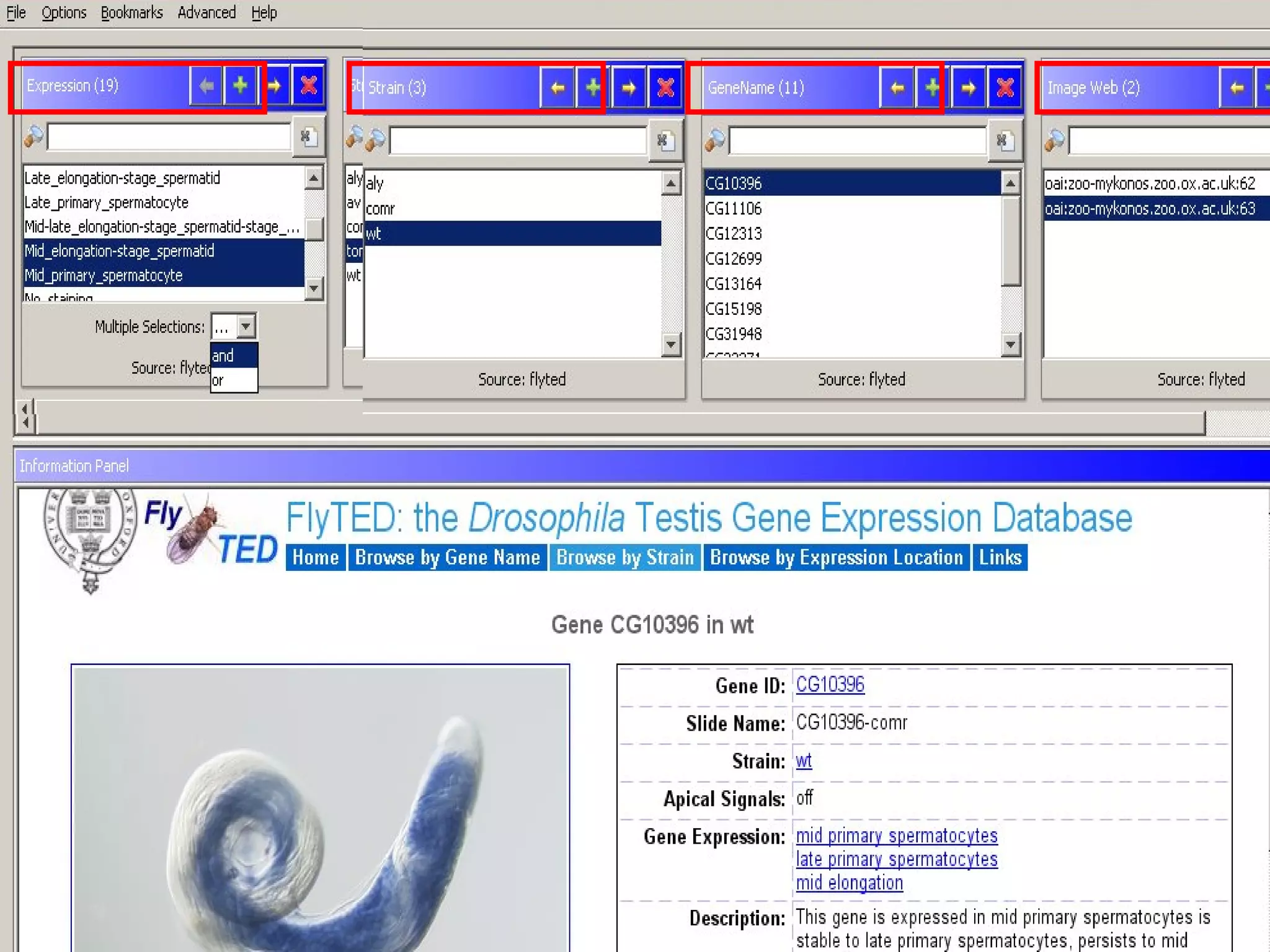
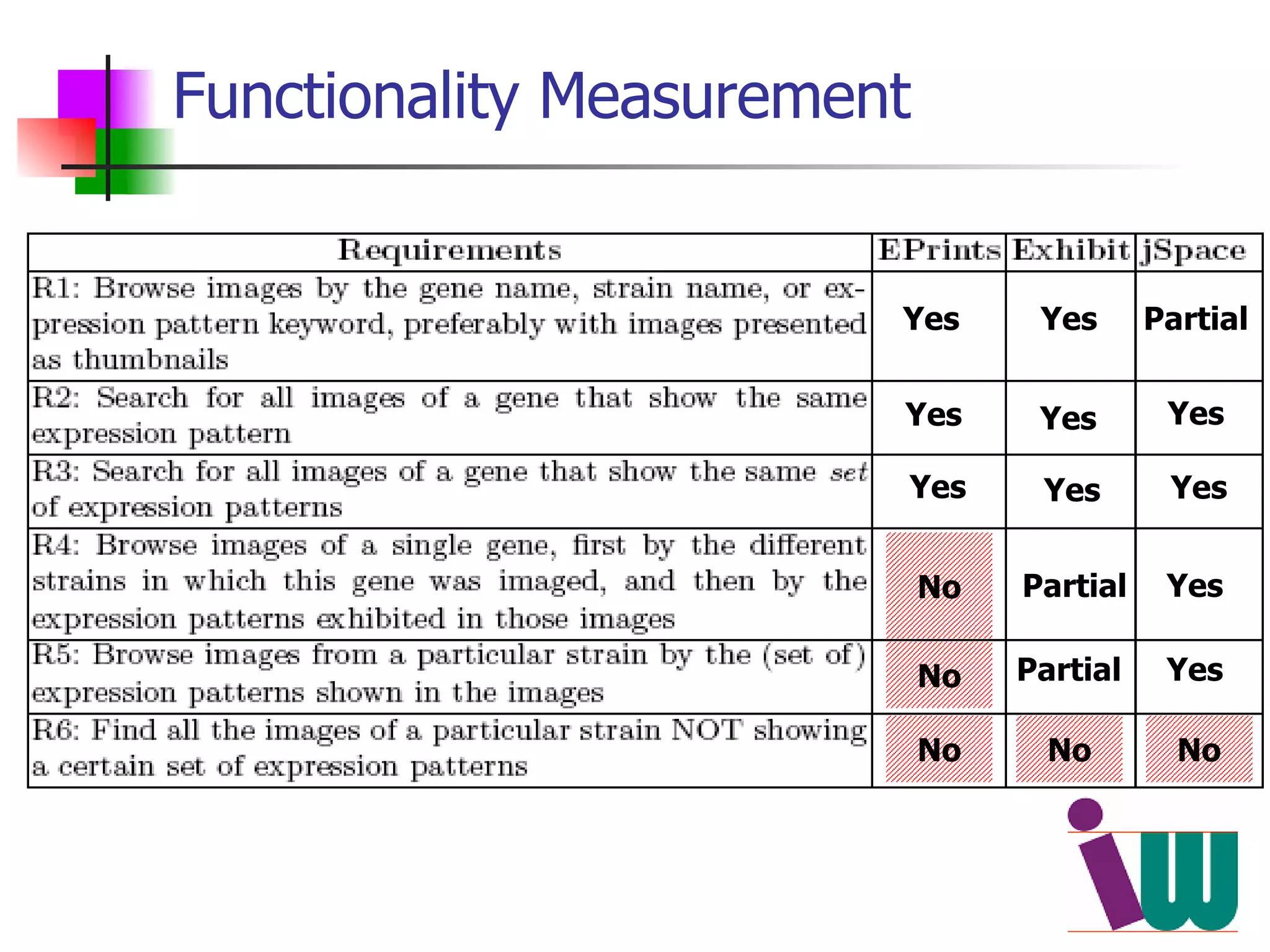
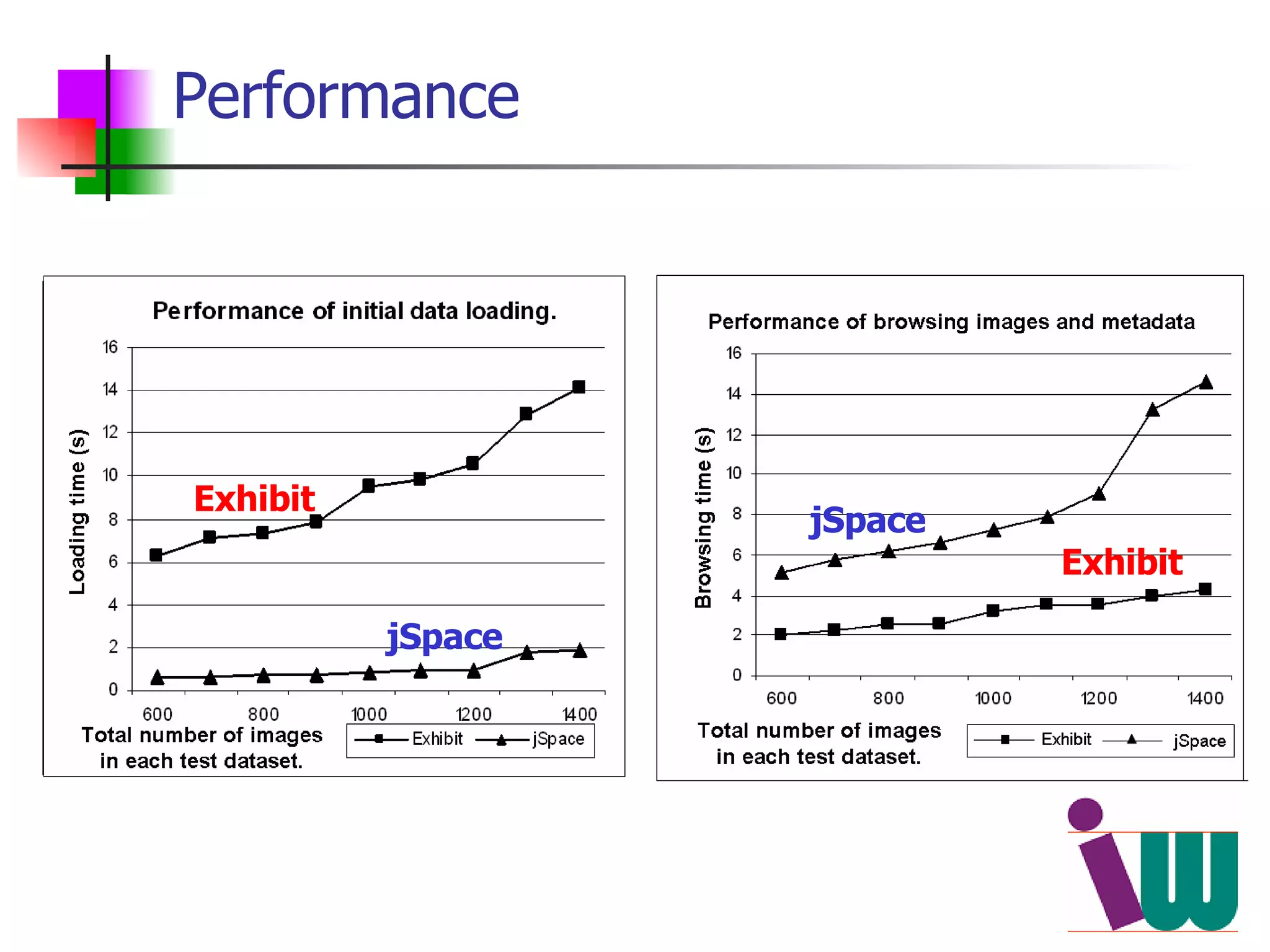
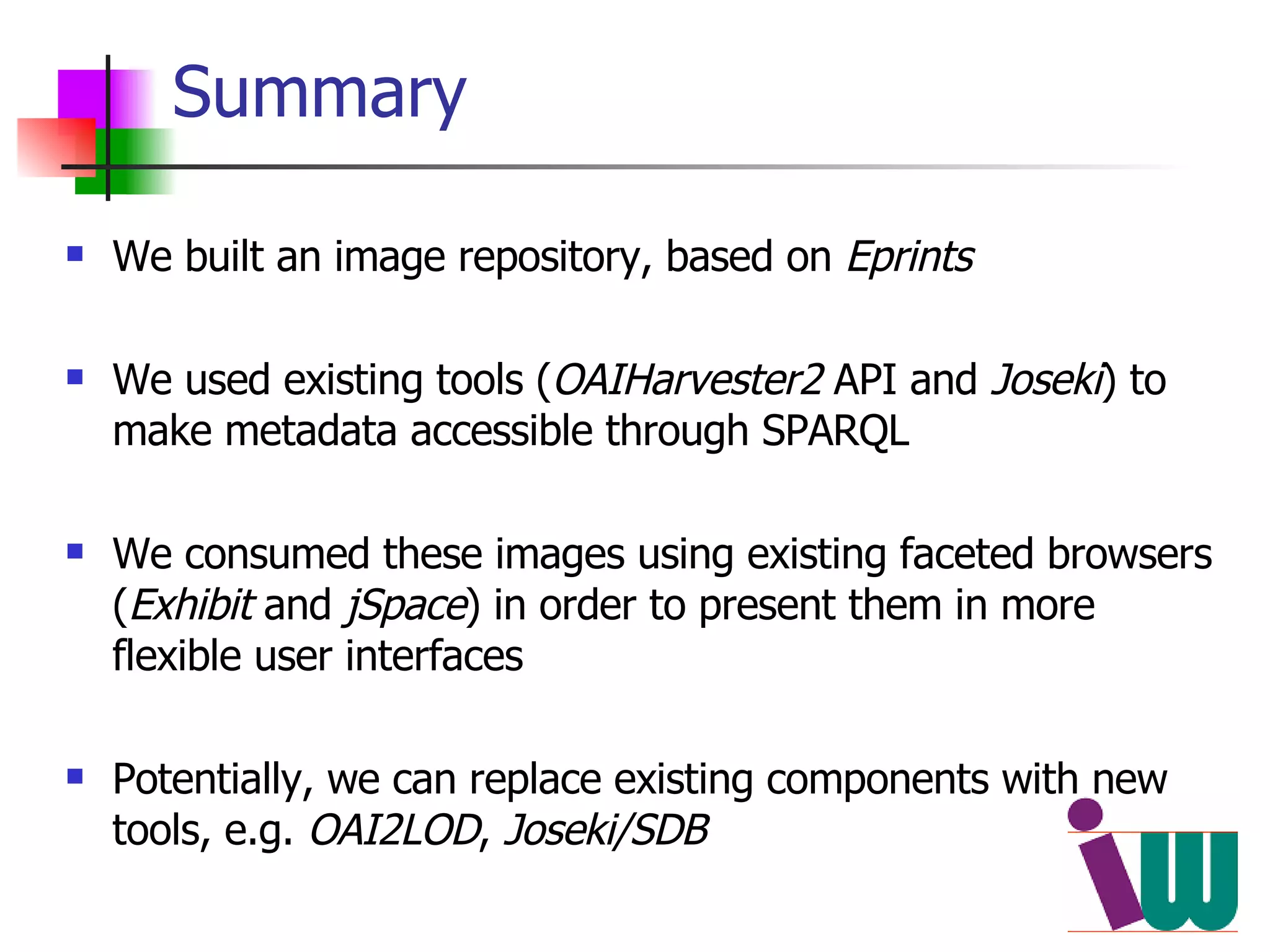
We built a semantic web image repository called FlyTED to publish Drosophila gene expression images online and make them searchable. We used existing tools like Eprints to store metadata and images and made the metadata accessible through SPARQL. We then consumed the images and metadata in faceted browsers like Exhibit and jSpace to present them in more flexible user interfaces. This allows components like the metadata storage to potentially be replaced in the future with new tools.
![Building a Semantic Web Image Repository for Biological Research Images Jun Zhao , Graham Klyne and David Shotton [email_address] Image Bioinformatics Research Group Department of Zoology University of Oxford, UK](https://image.slidesharecdn.com/2008junzhaoeswc-1213209032733549-8/75/2008-Jun-Zhao-Eswc-1-2048.jpg)