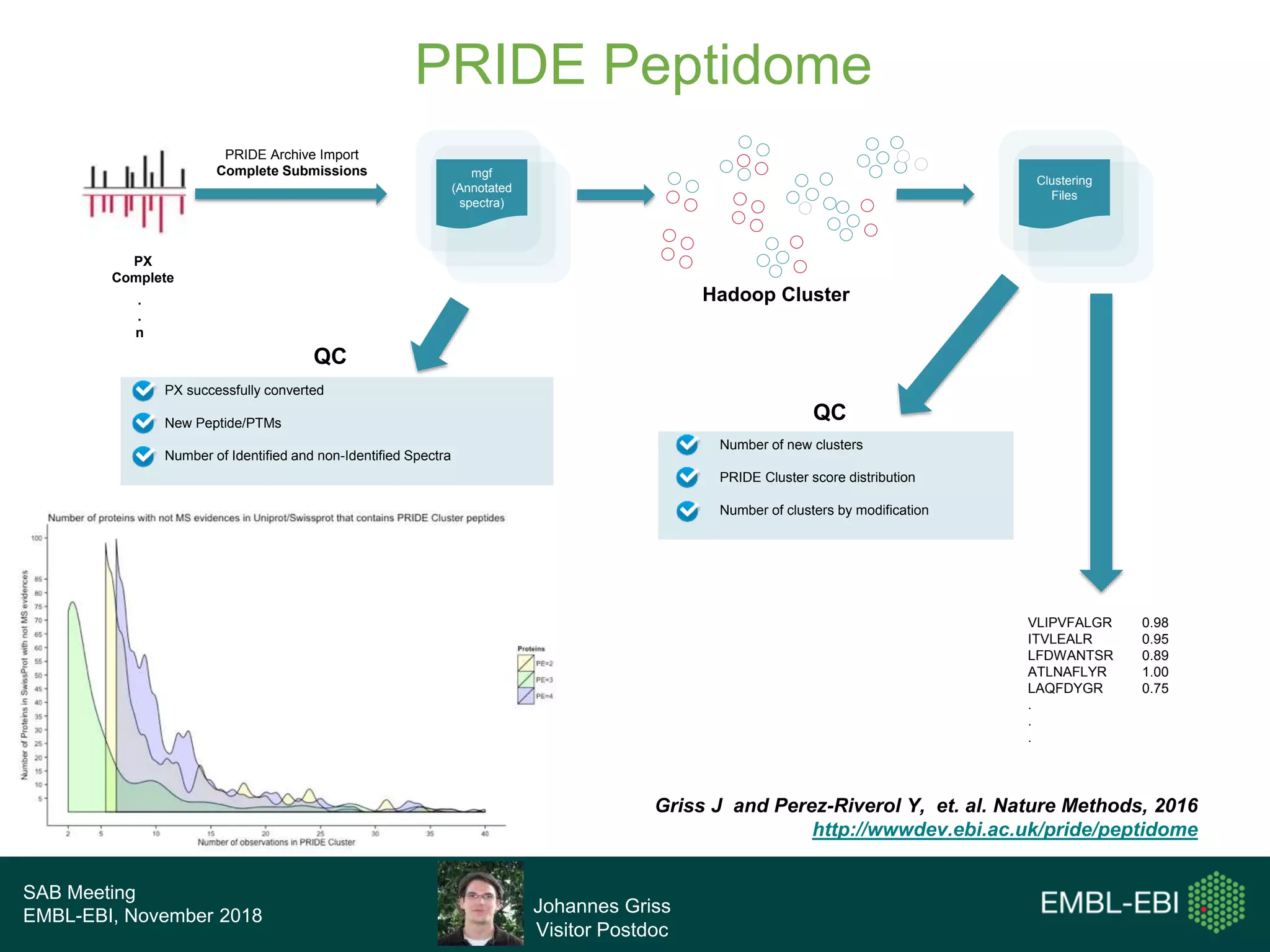
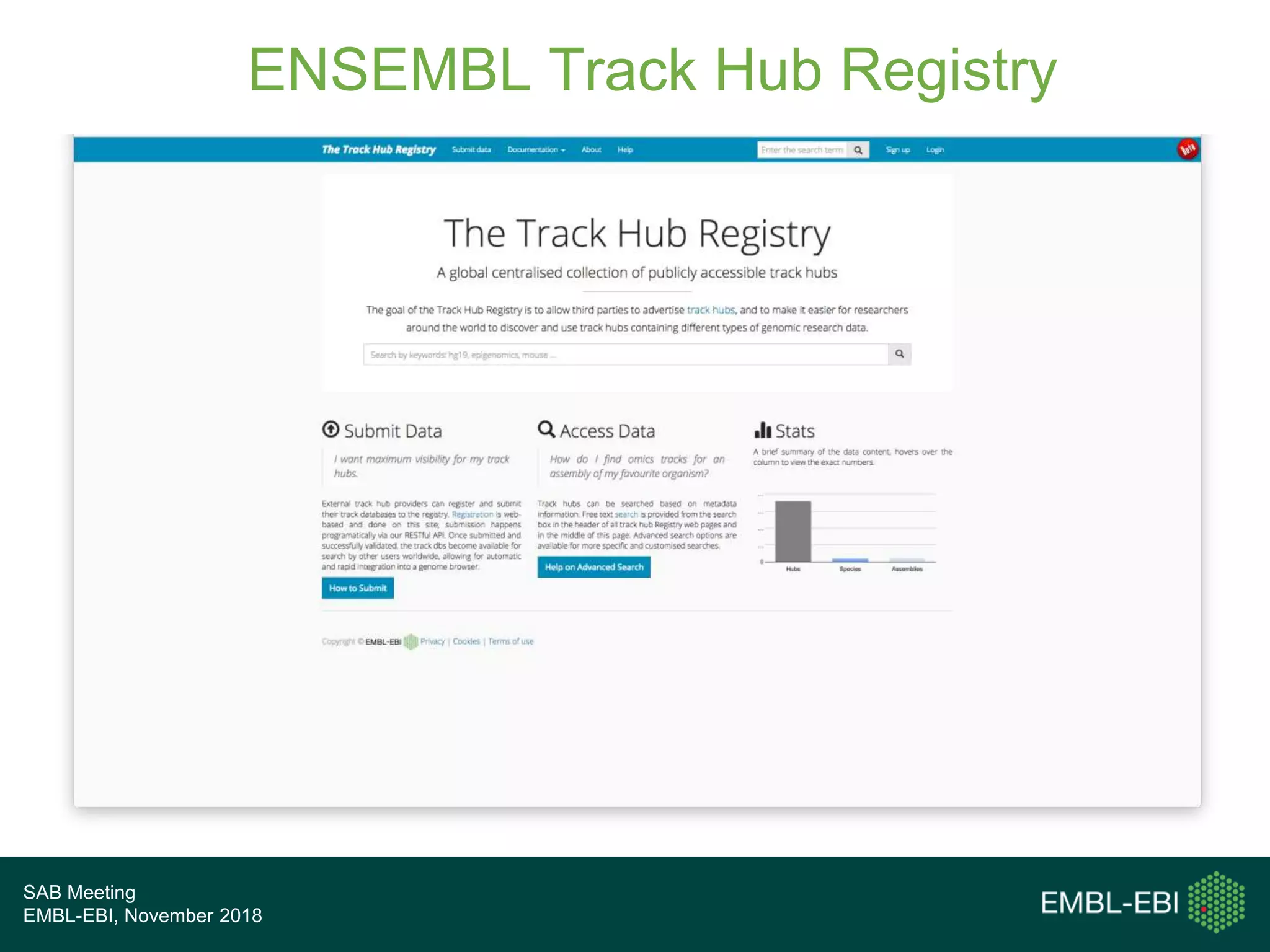
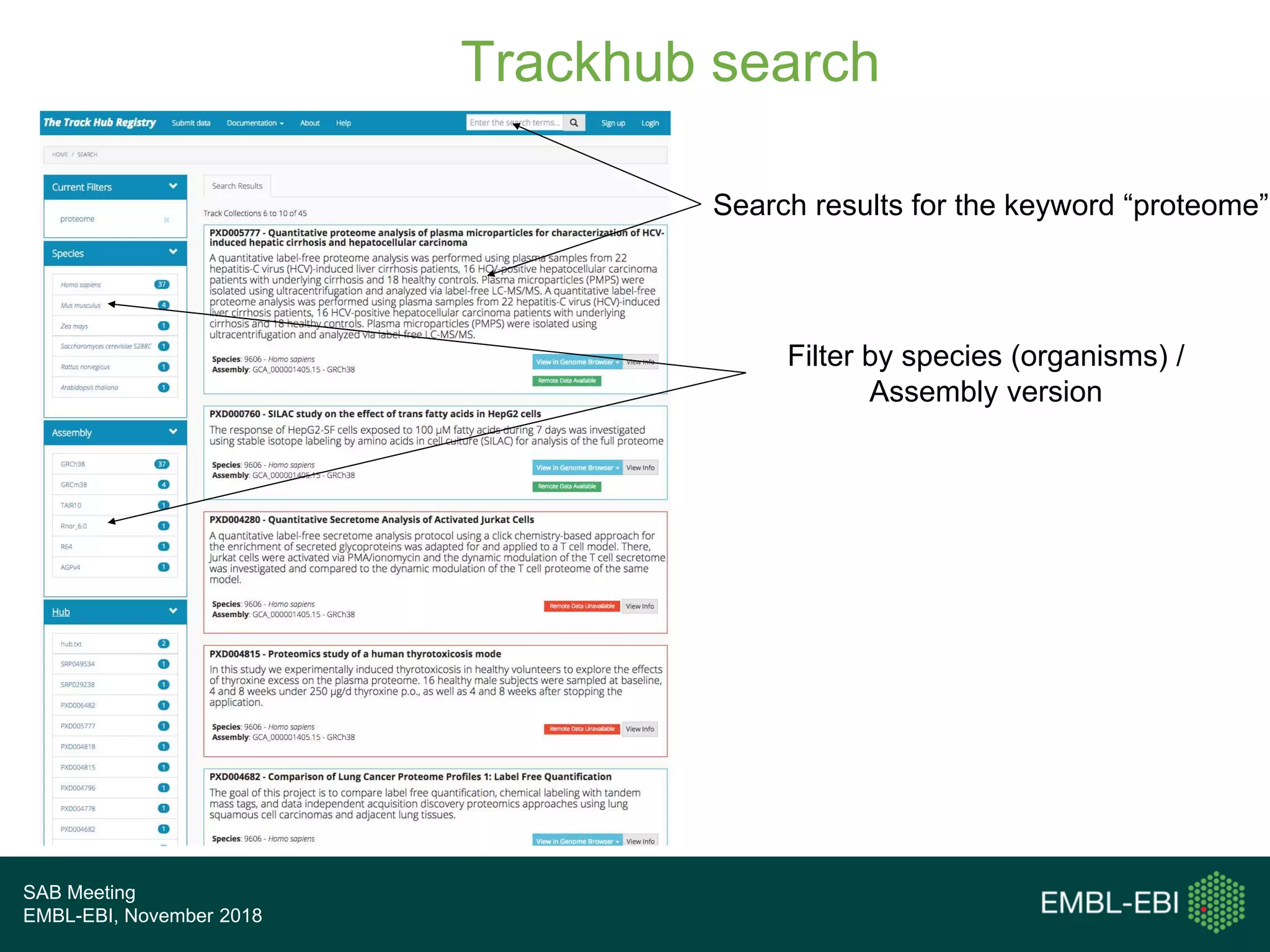
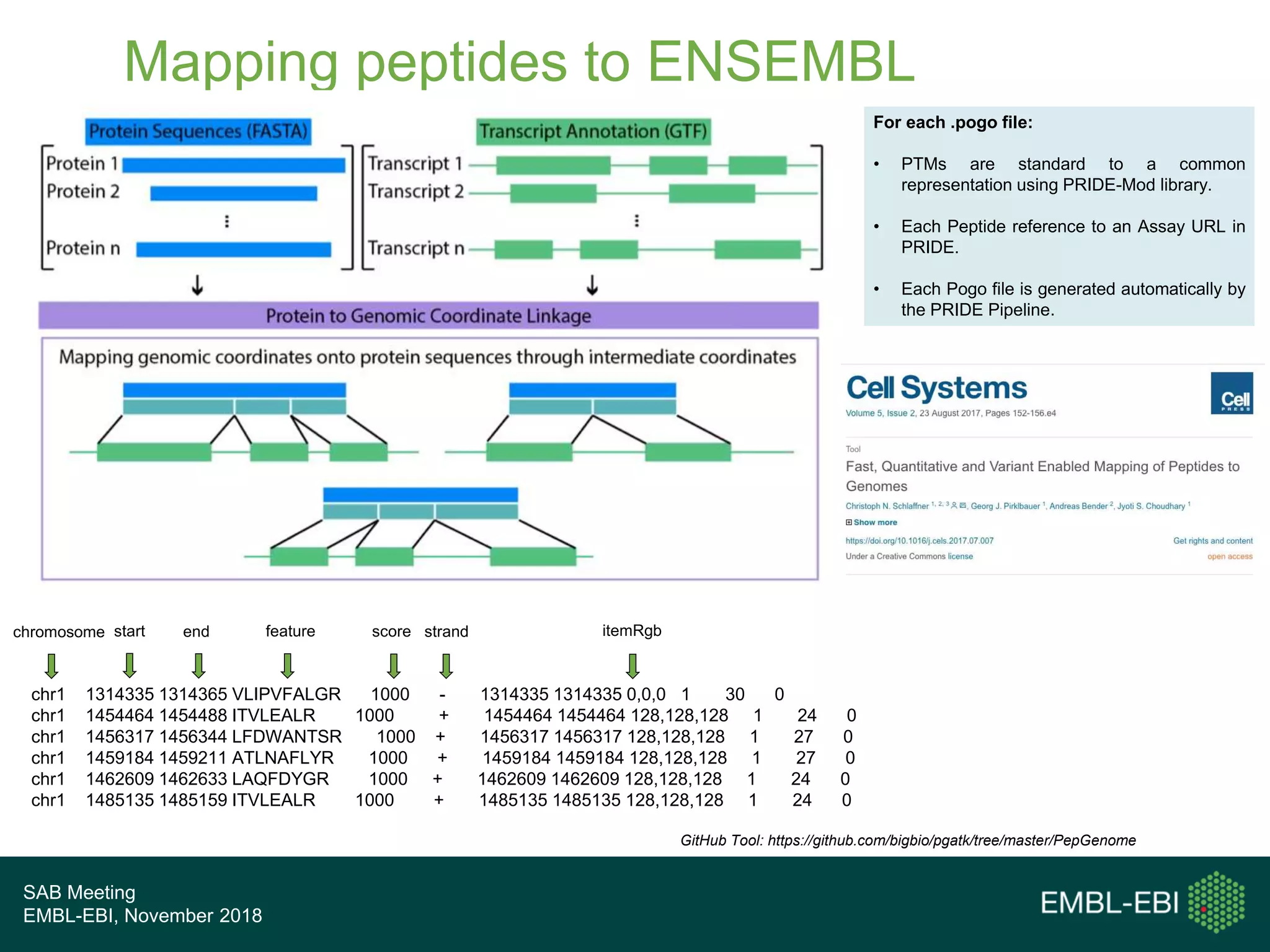
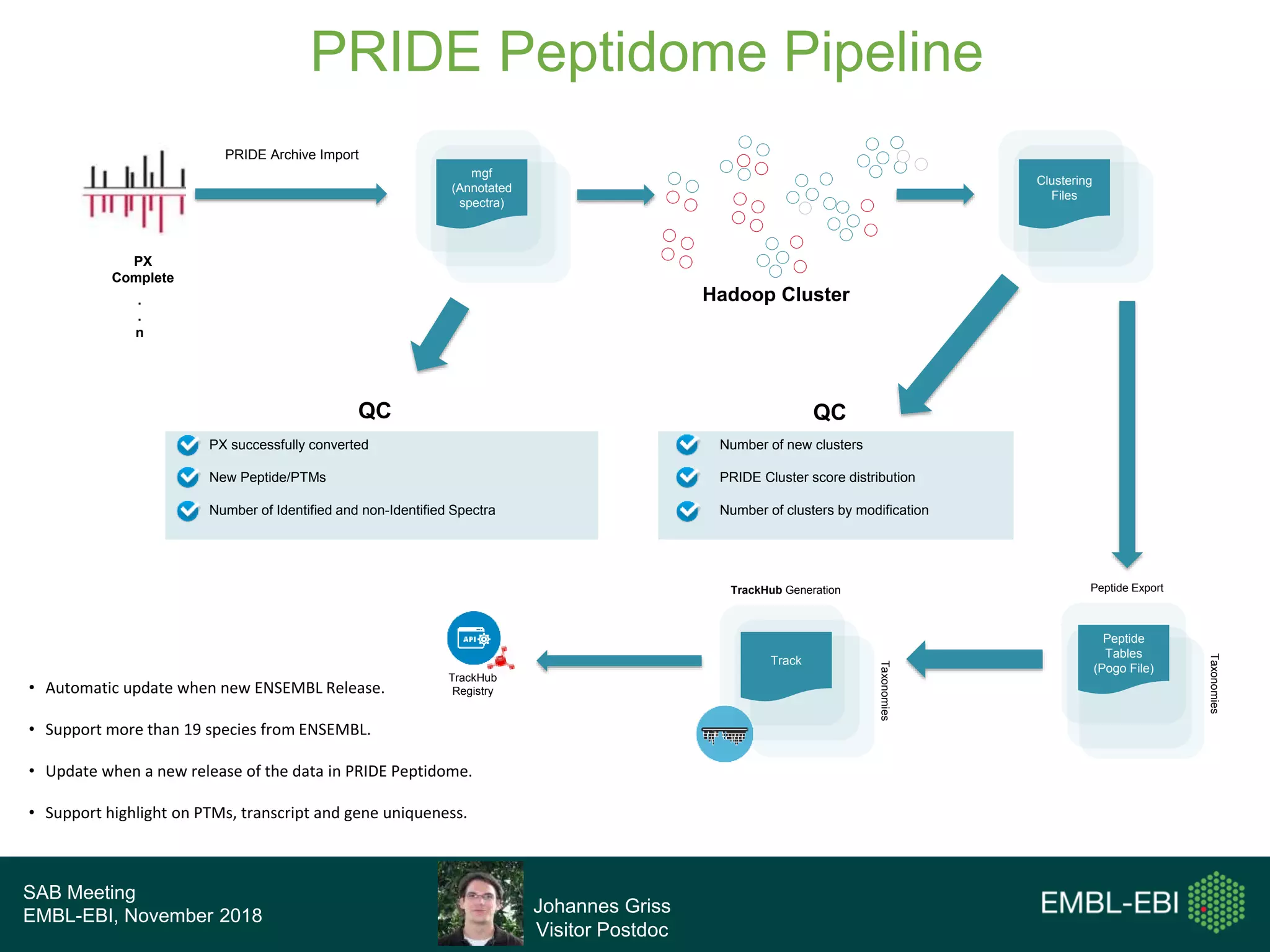
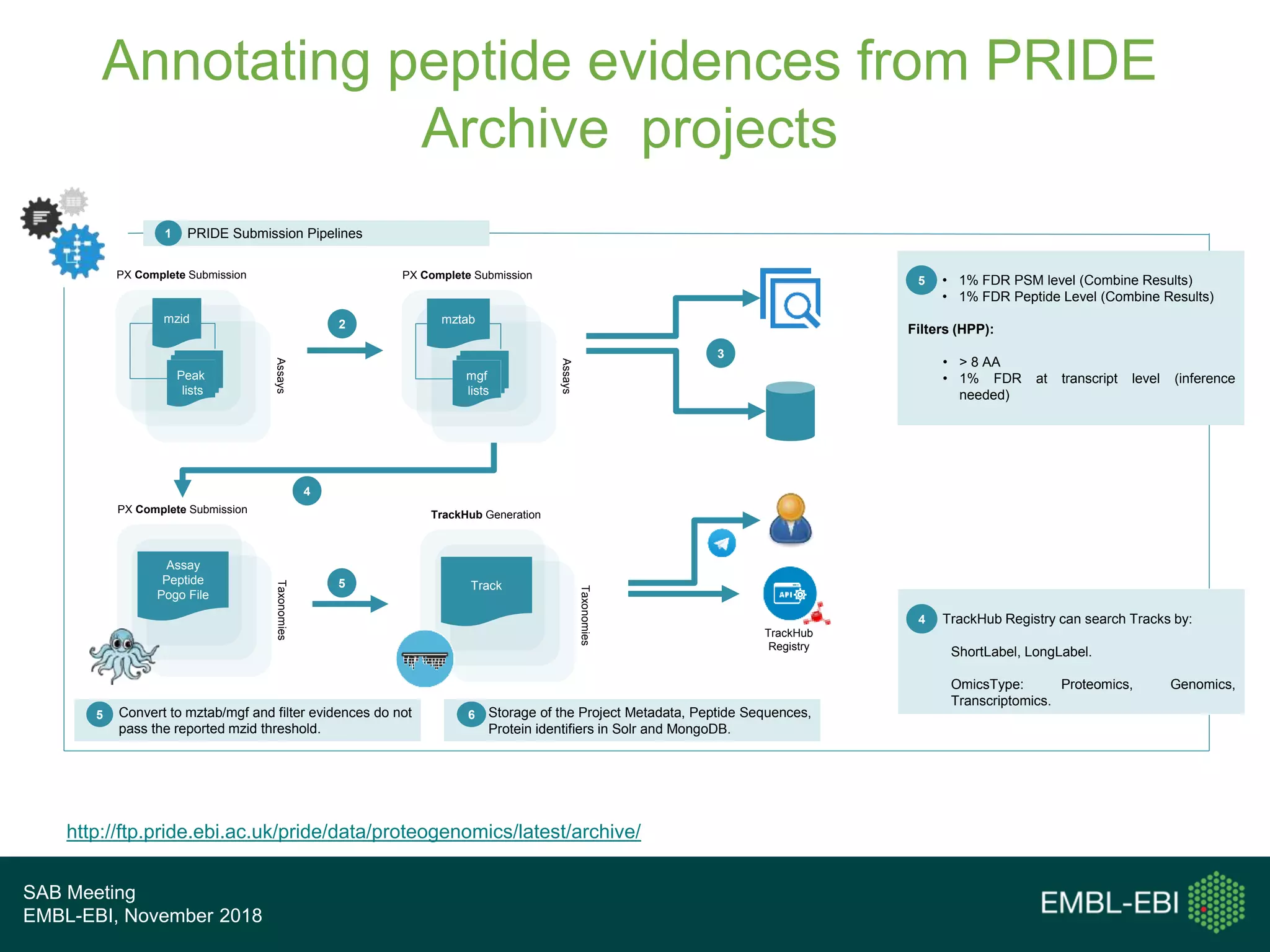
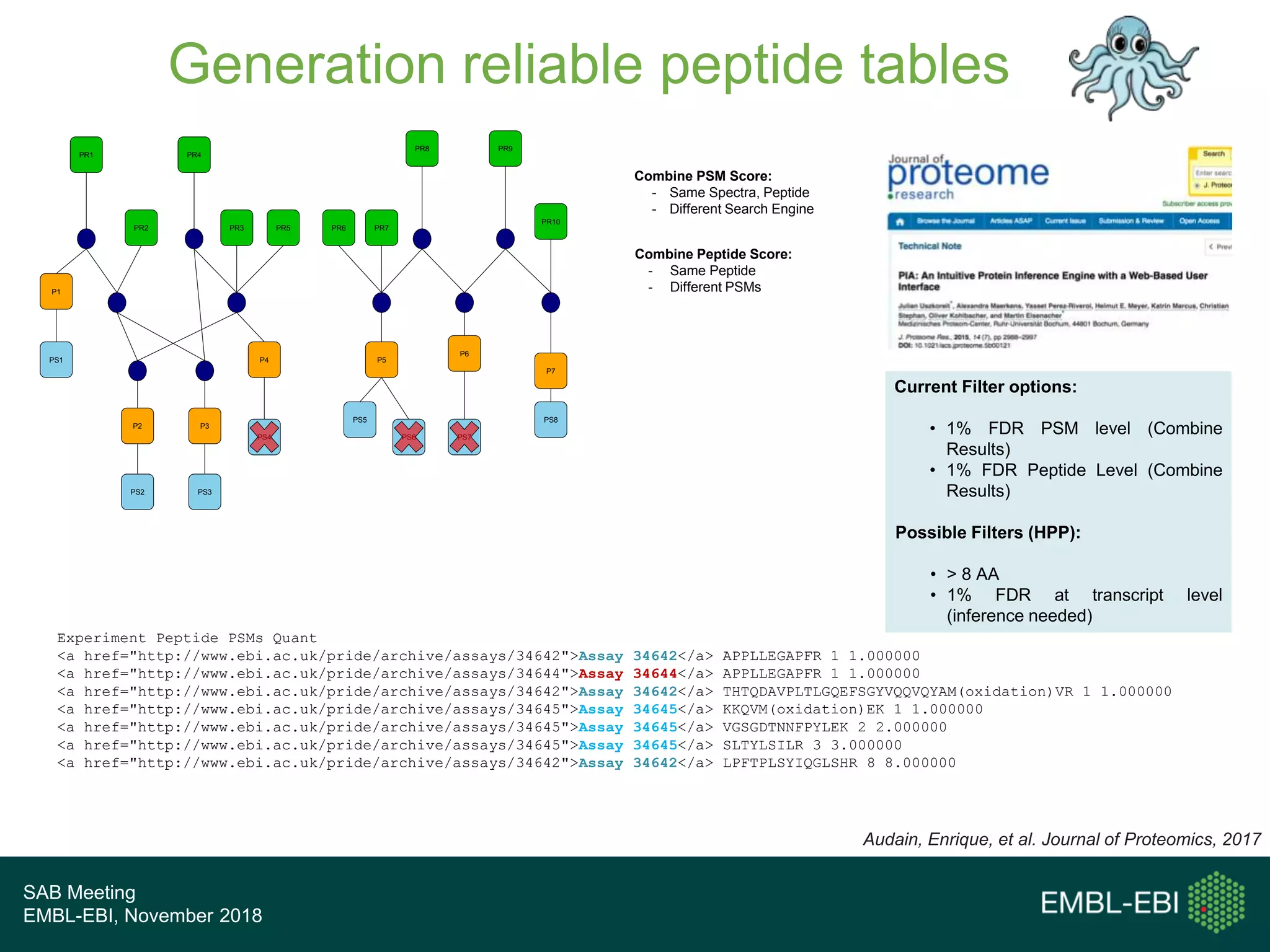
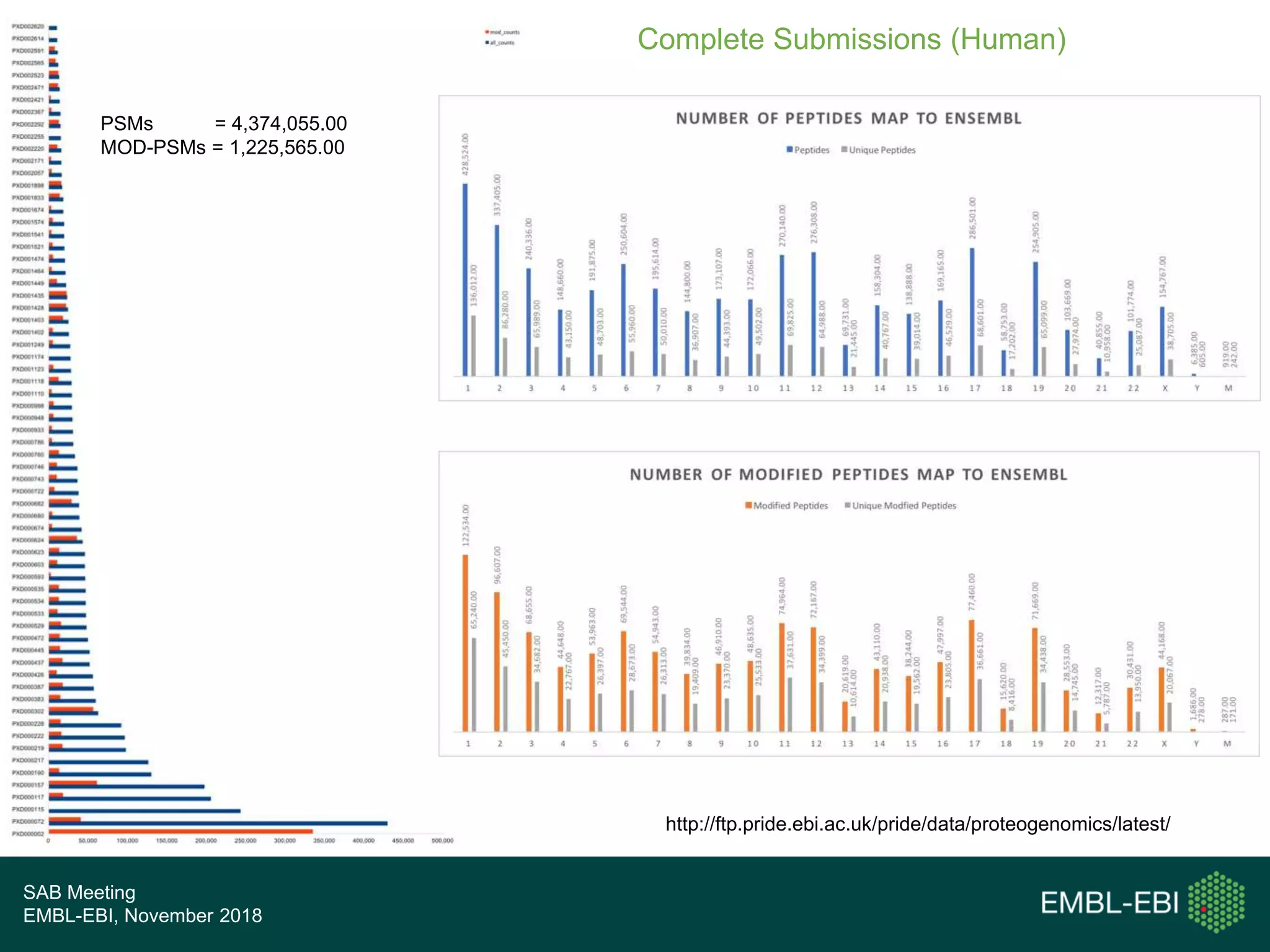
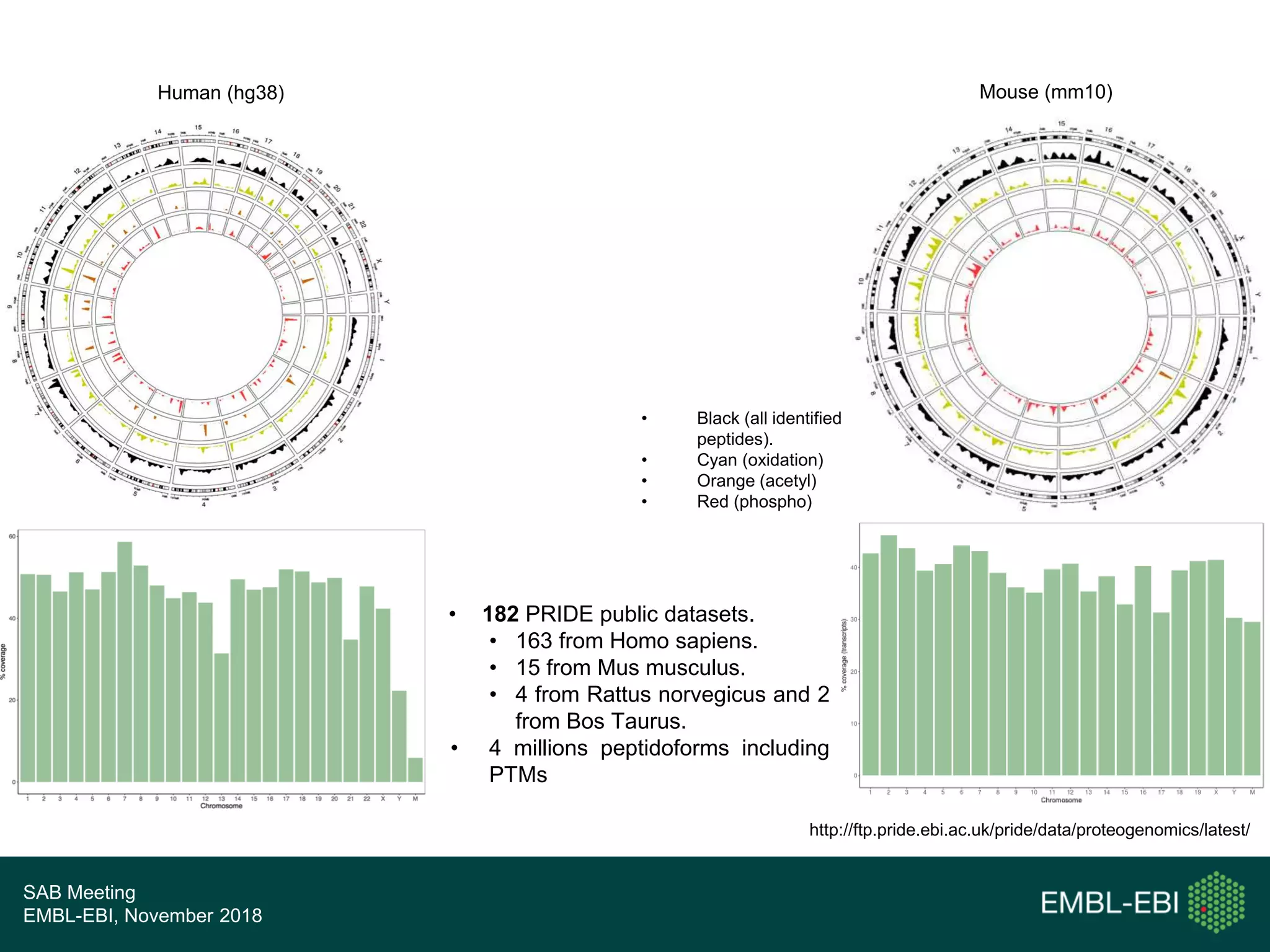
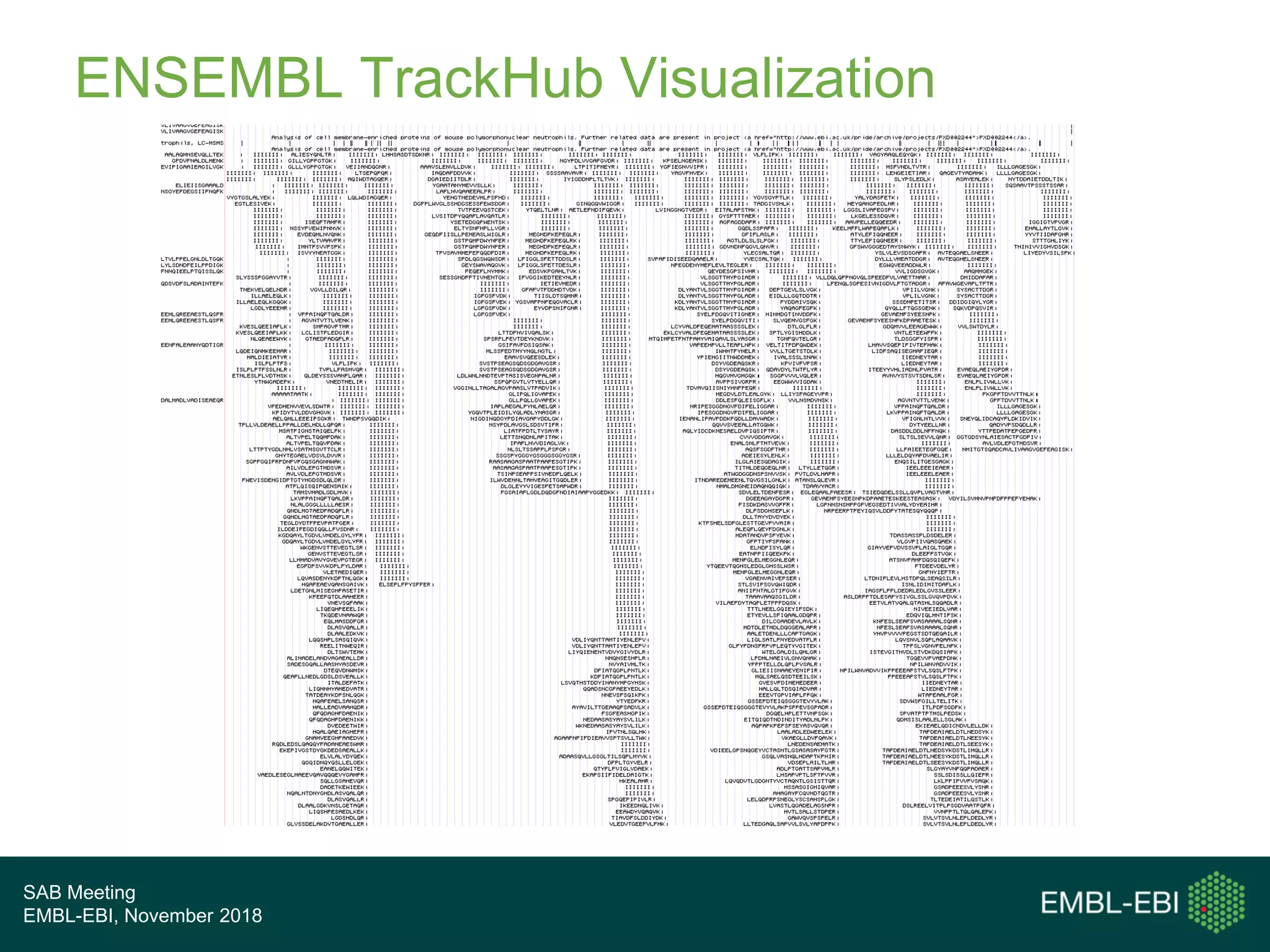
The PRIDE Resource Team is moving millions of peptide evidences from PRIDE submissions into EBI protein resources to provide a genomics context. This includes mapping post-translational modifications and variants to ENSEMBL tracks. Peptides from PRIDE are clustered, mapped to genomes using a GitHub tool, and visualized in the ENSEMBL TrackHub registry. Over 4 million peptidoforms including modifications have been mapped from 182 public datasets for human and mouse. The team aims to increase the number of mapped submissions and provide more associated metadata to improve protein annotations.