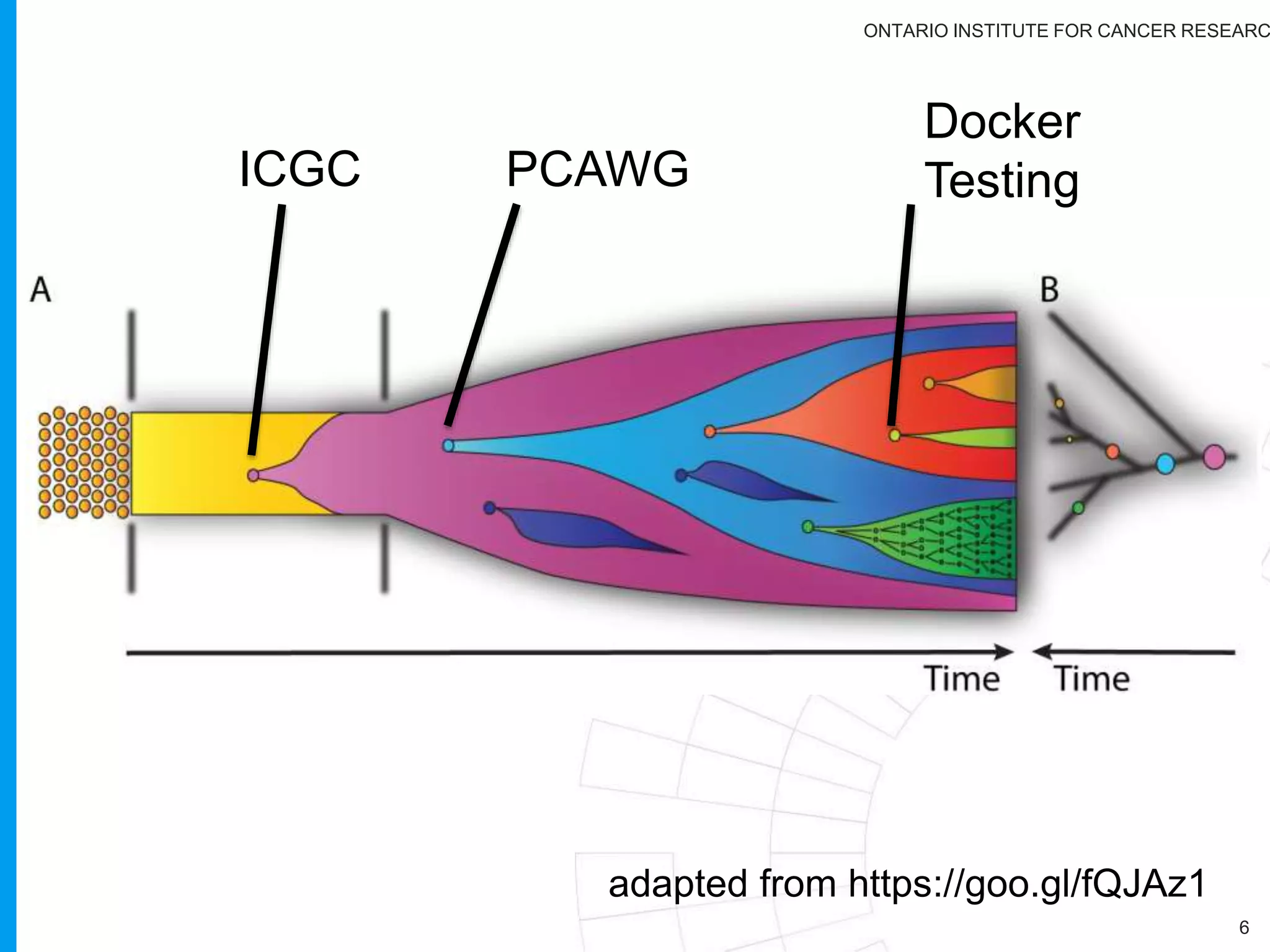
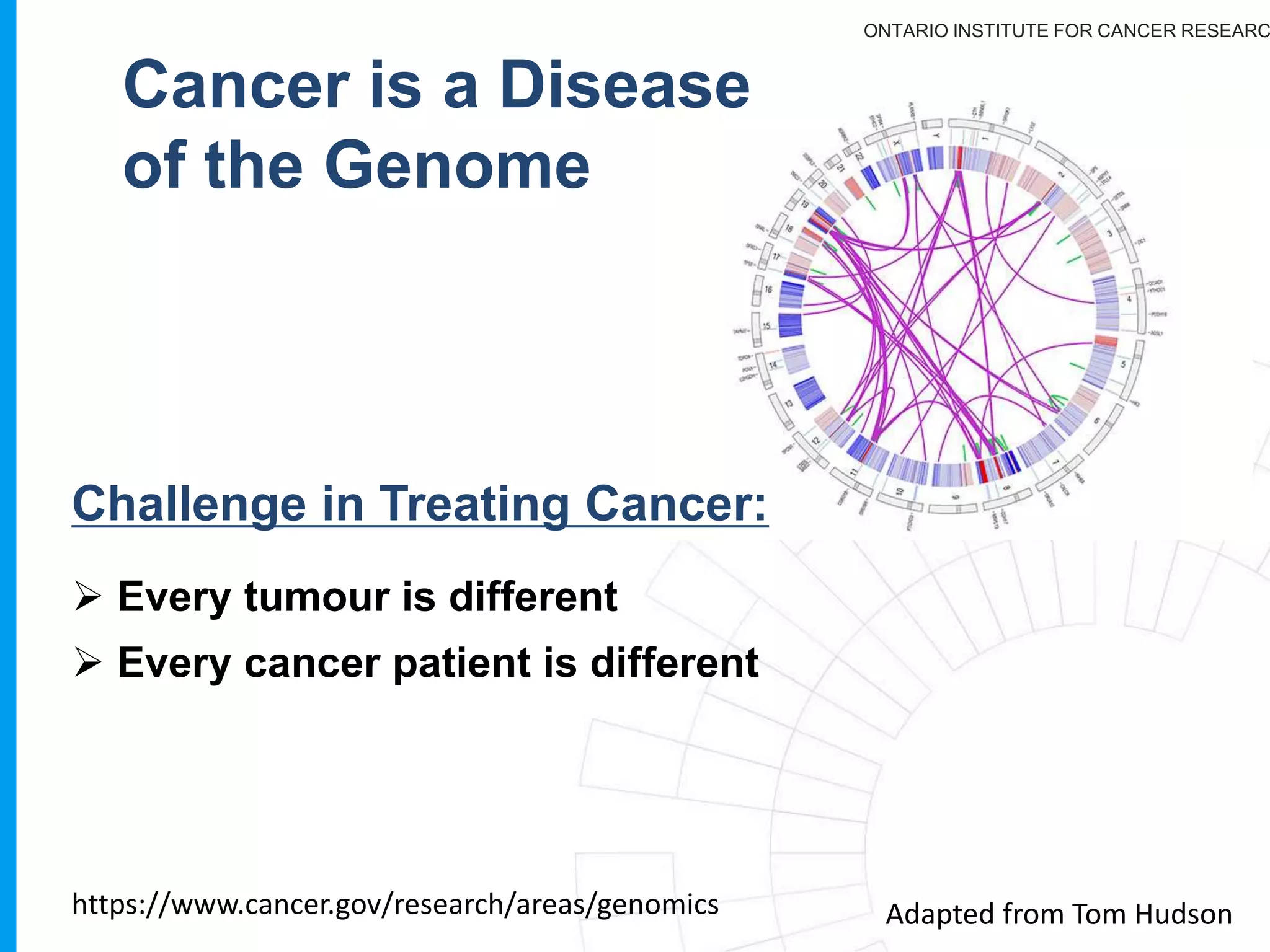
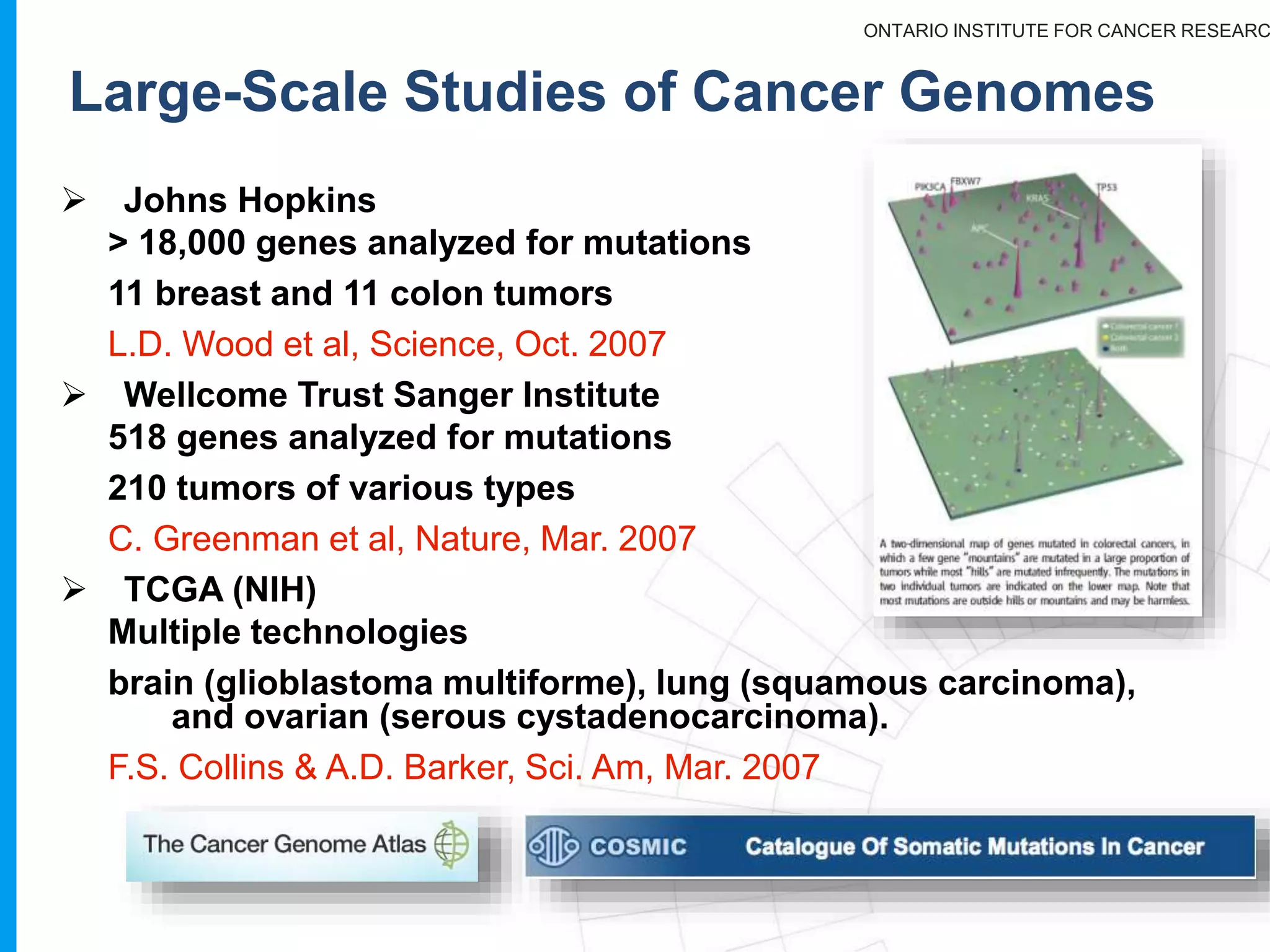
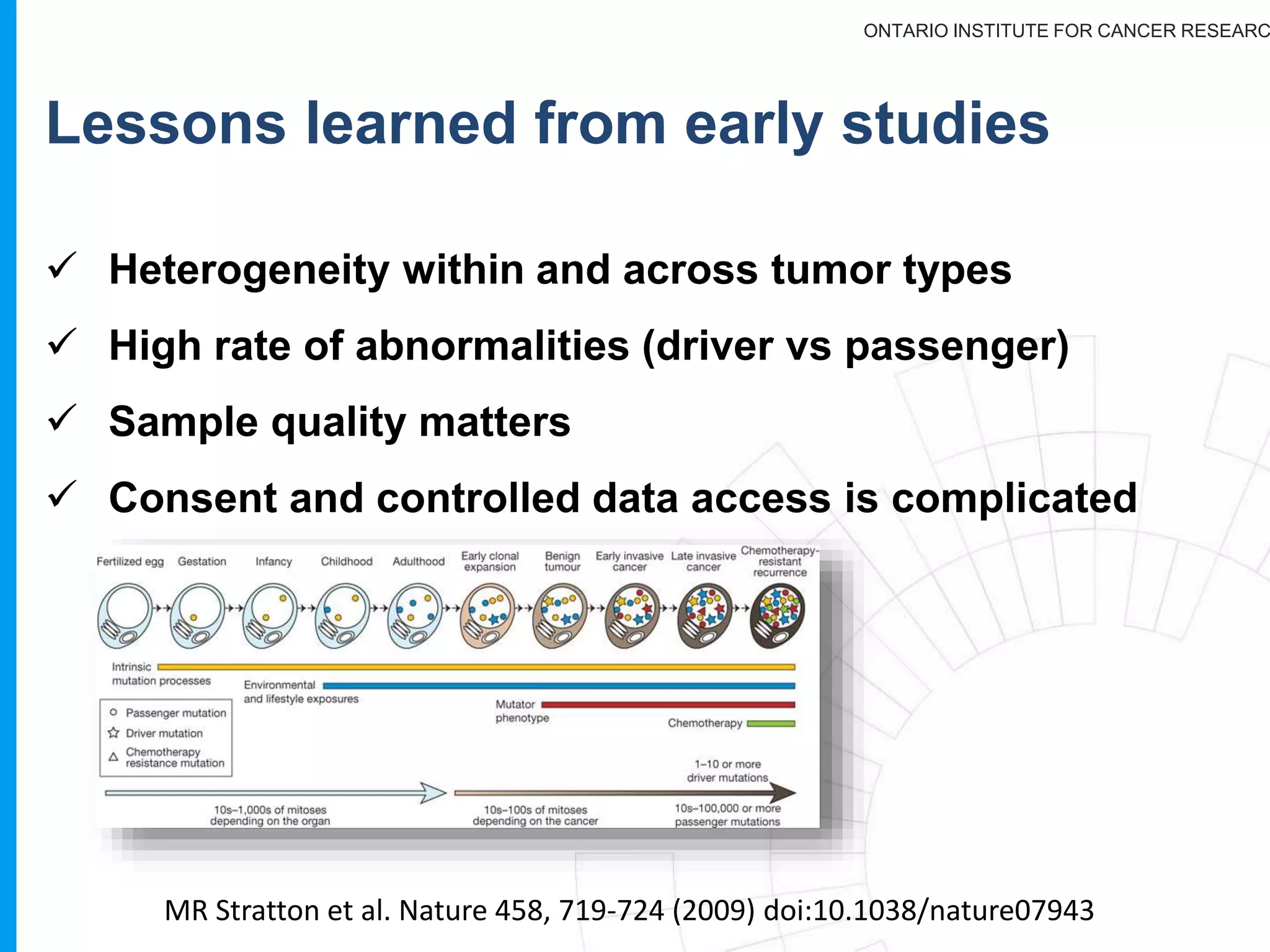
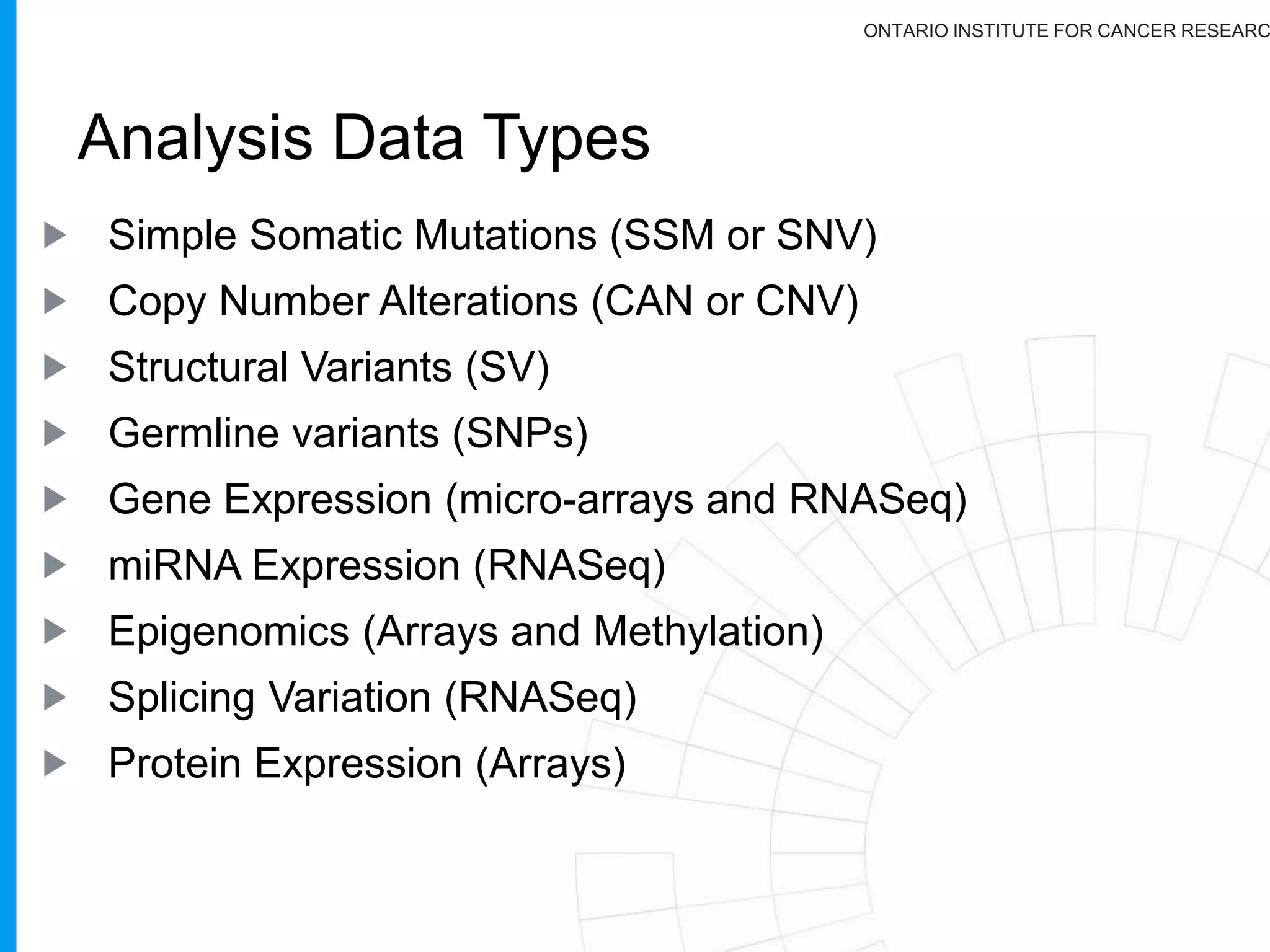
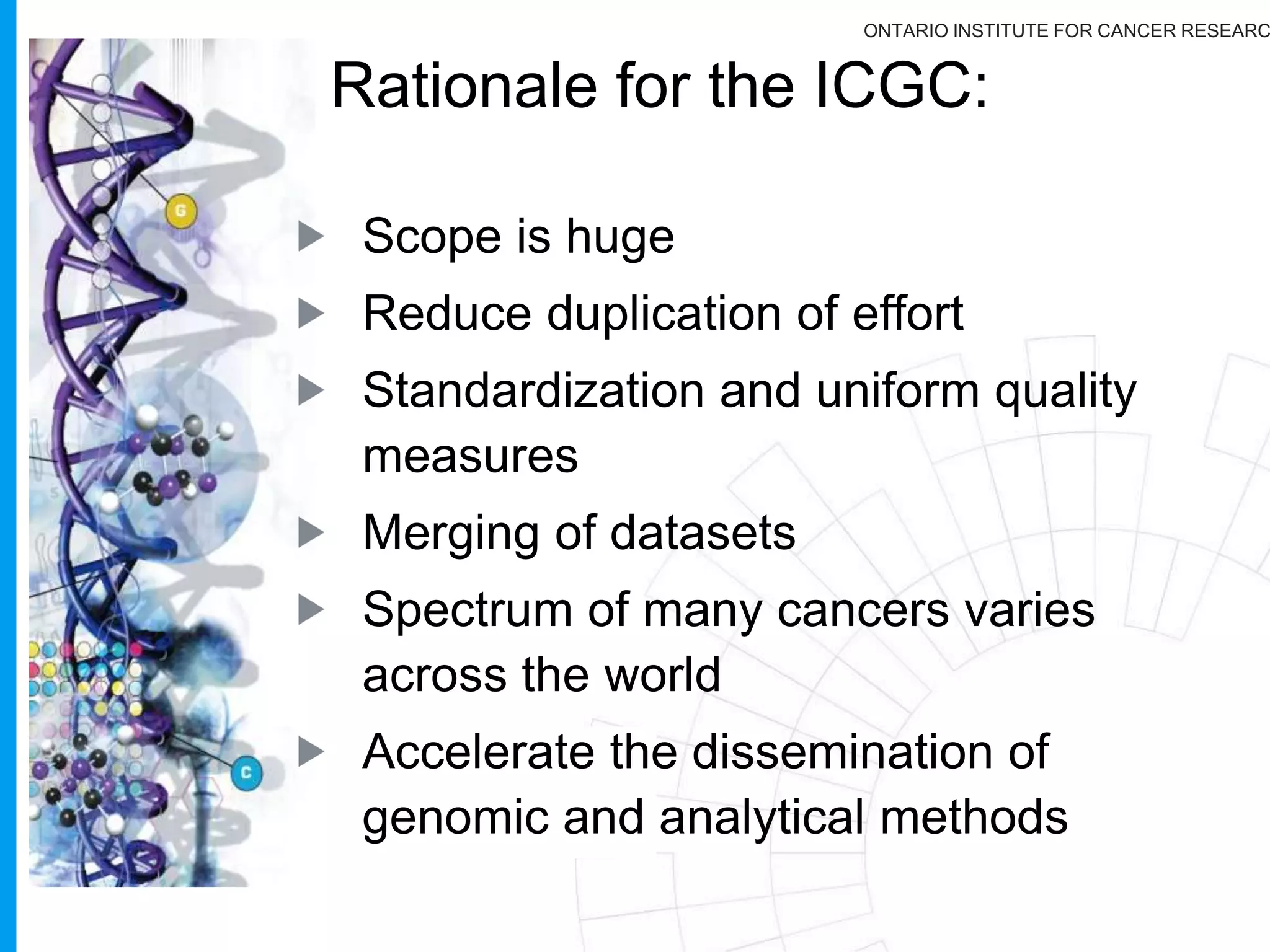
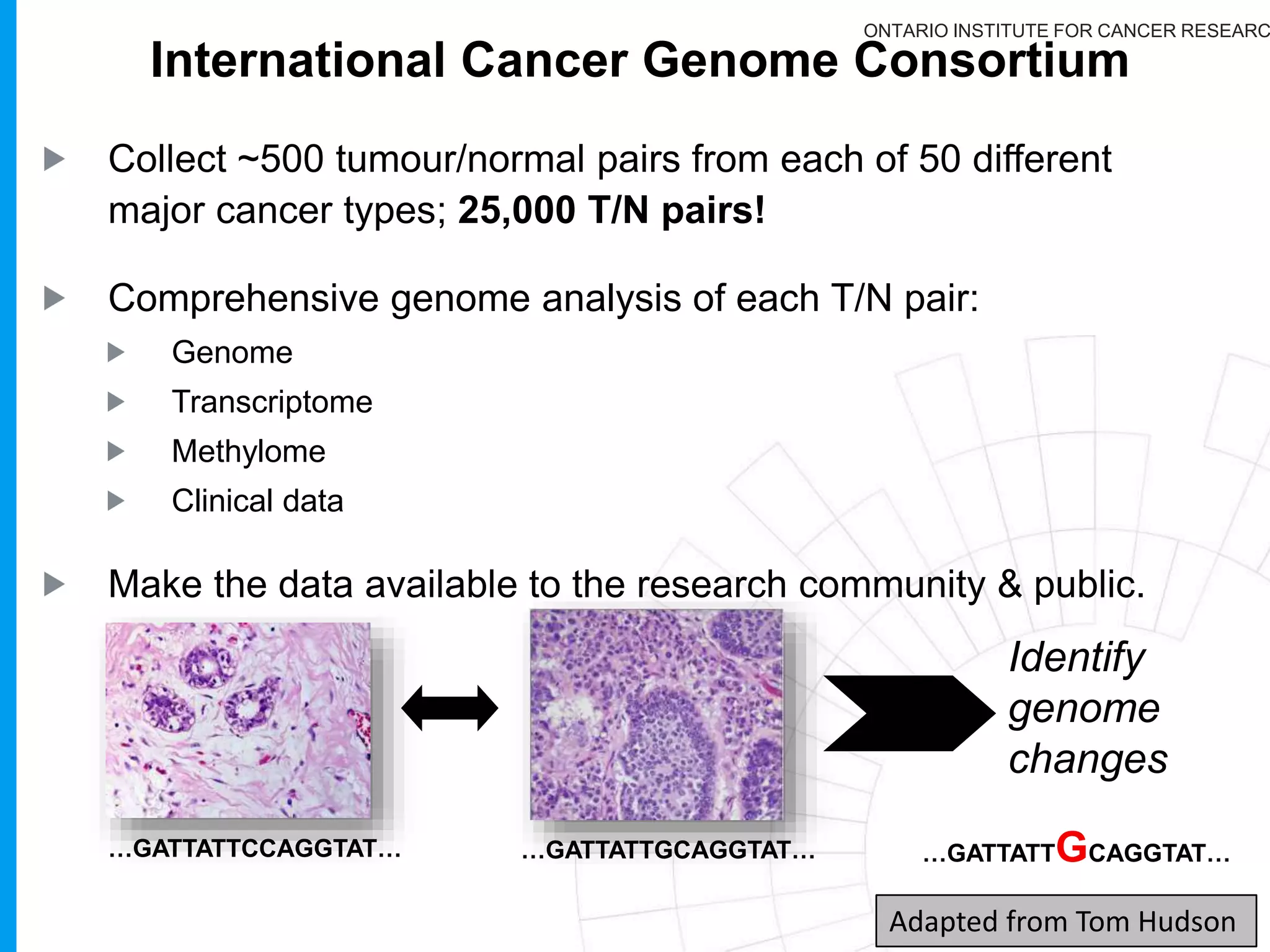
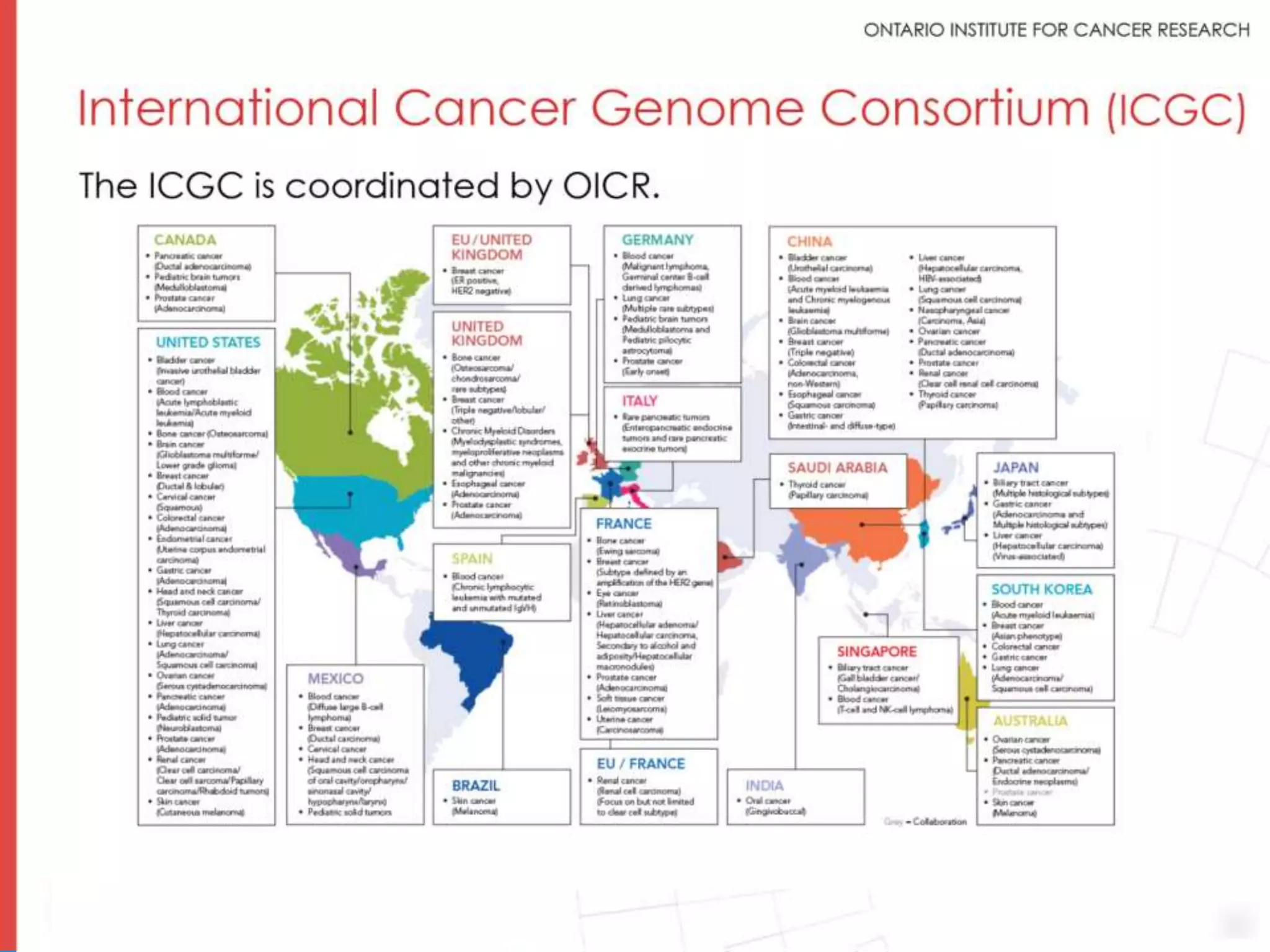
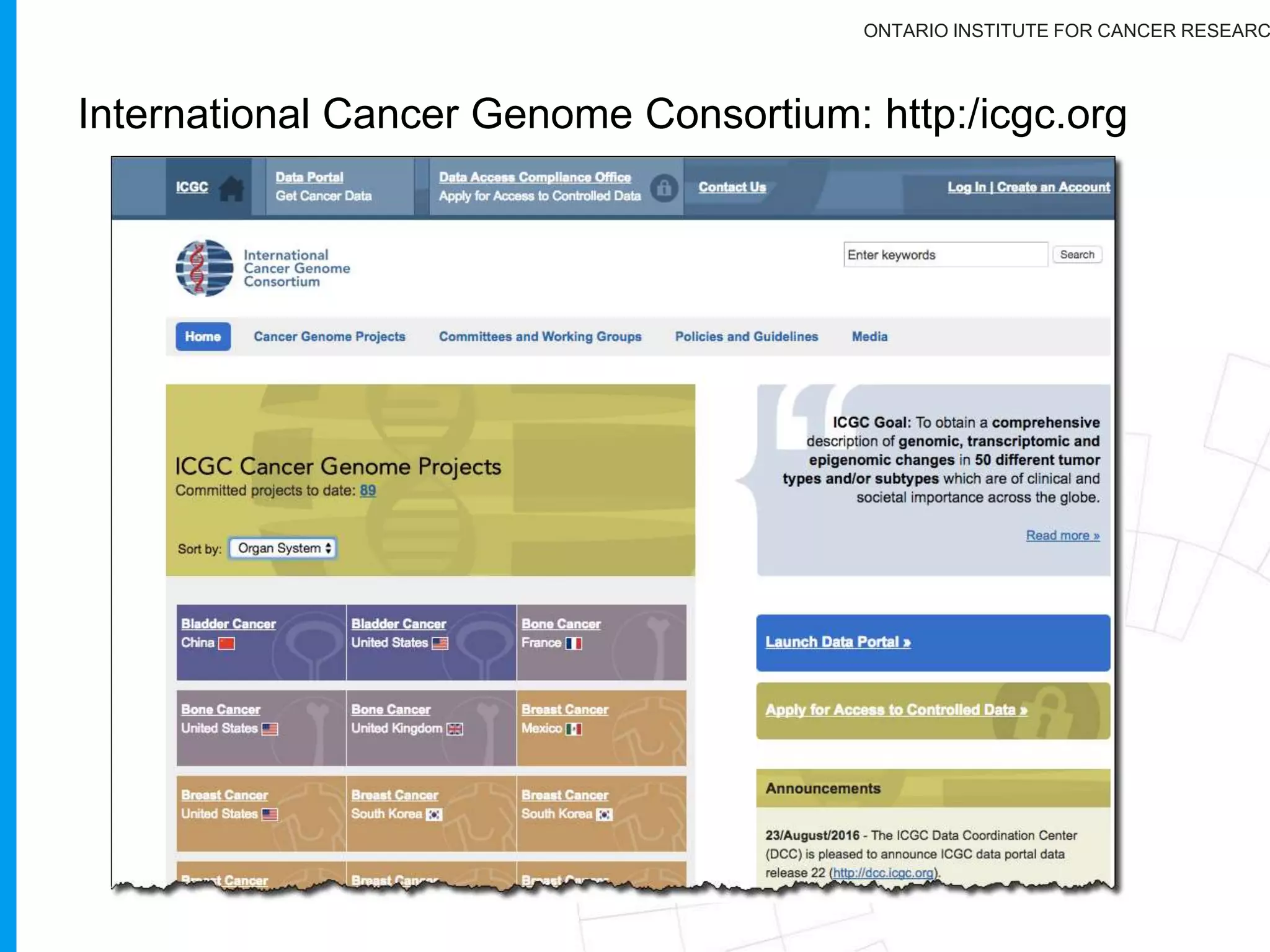
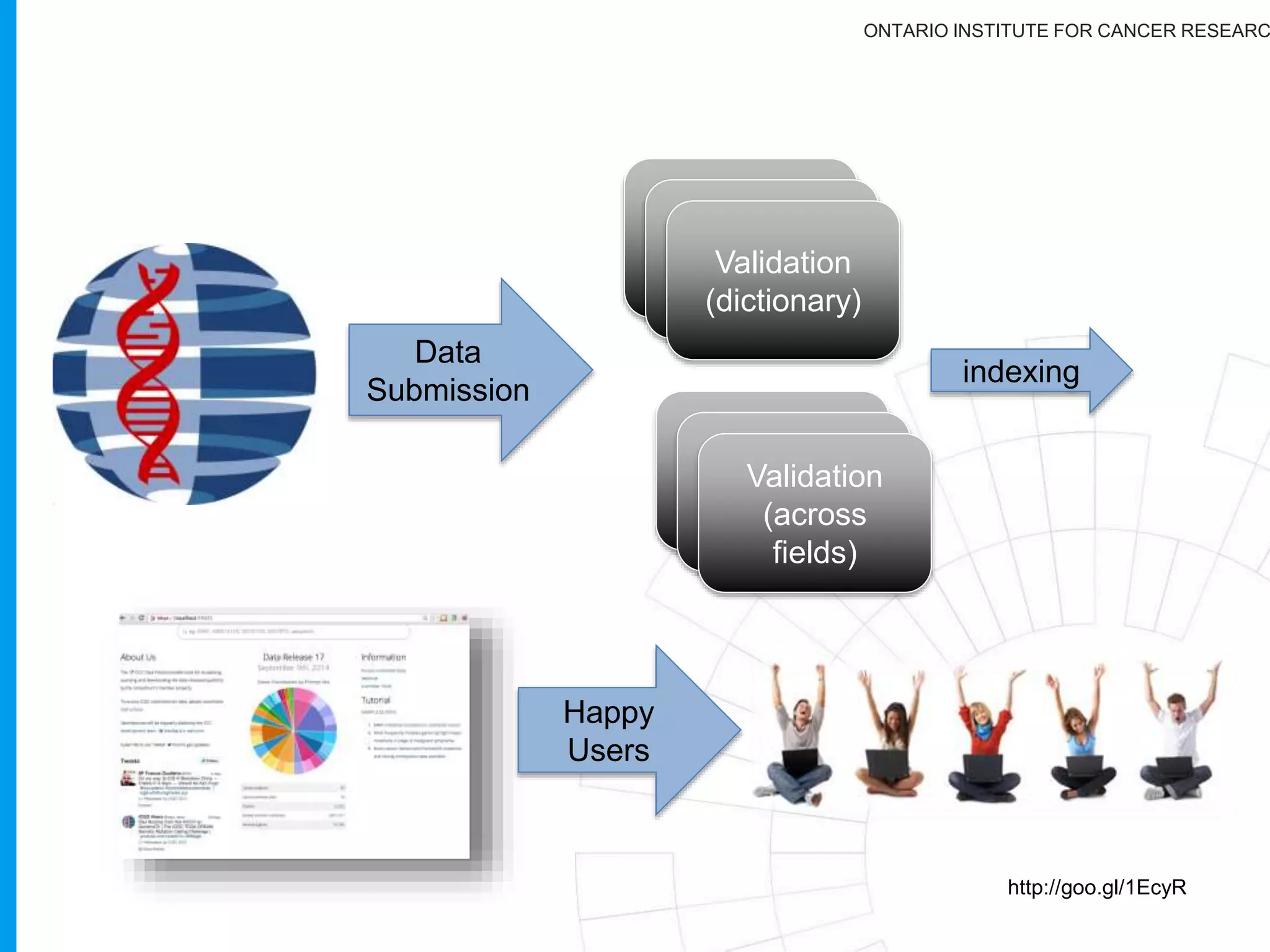
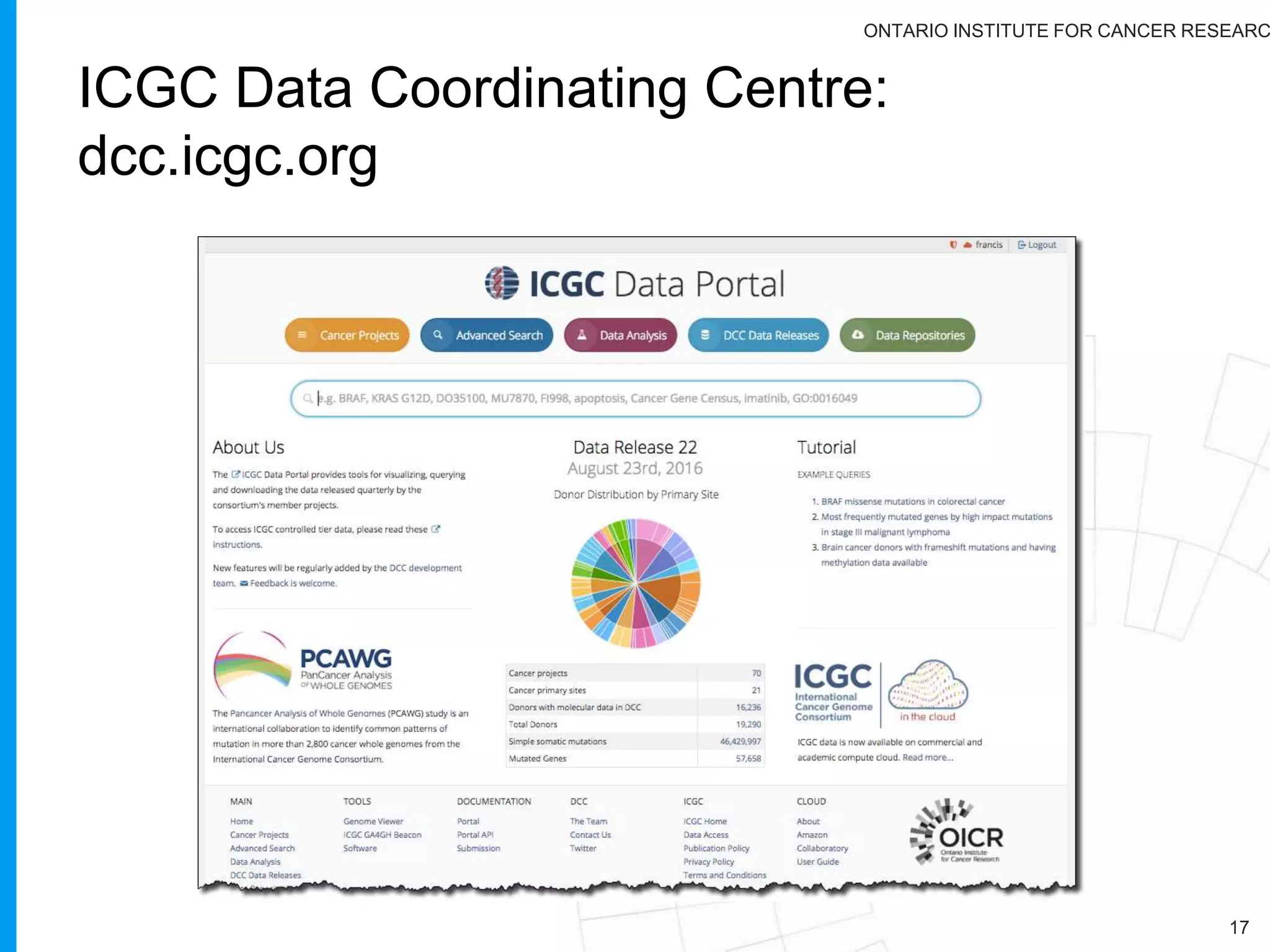
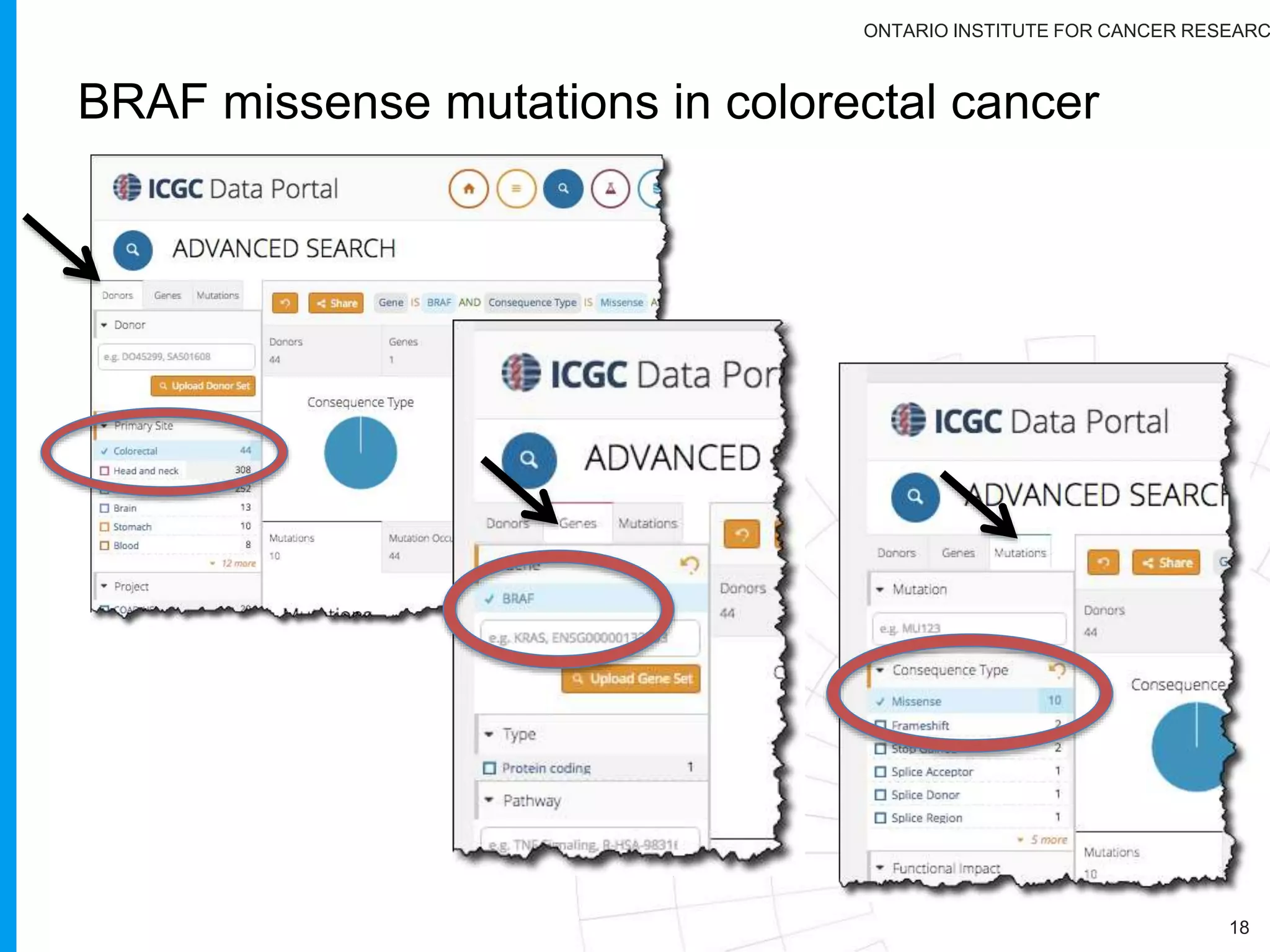
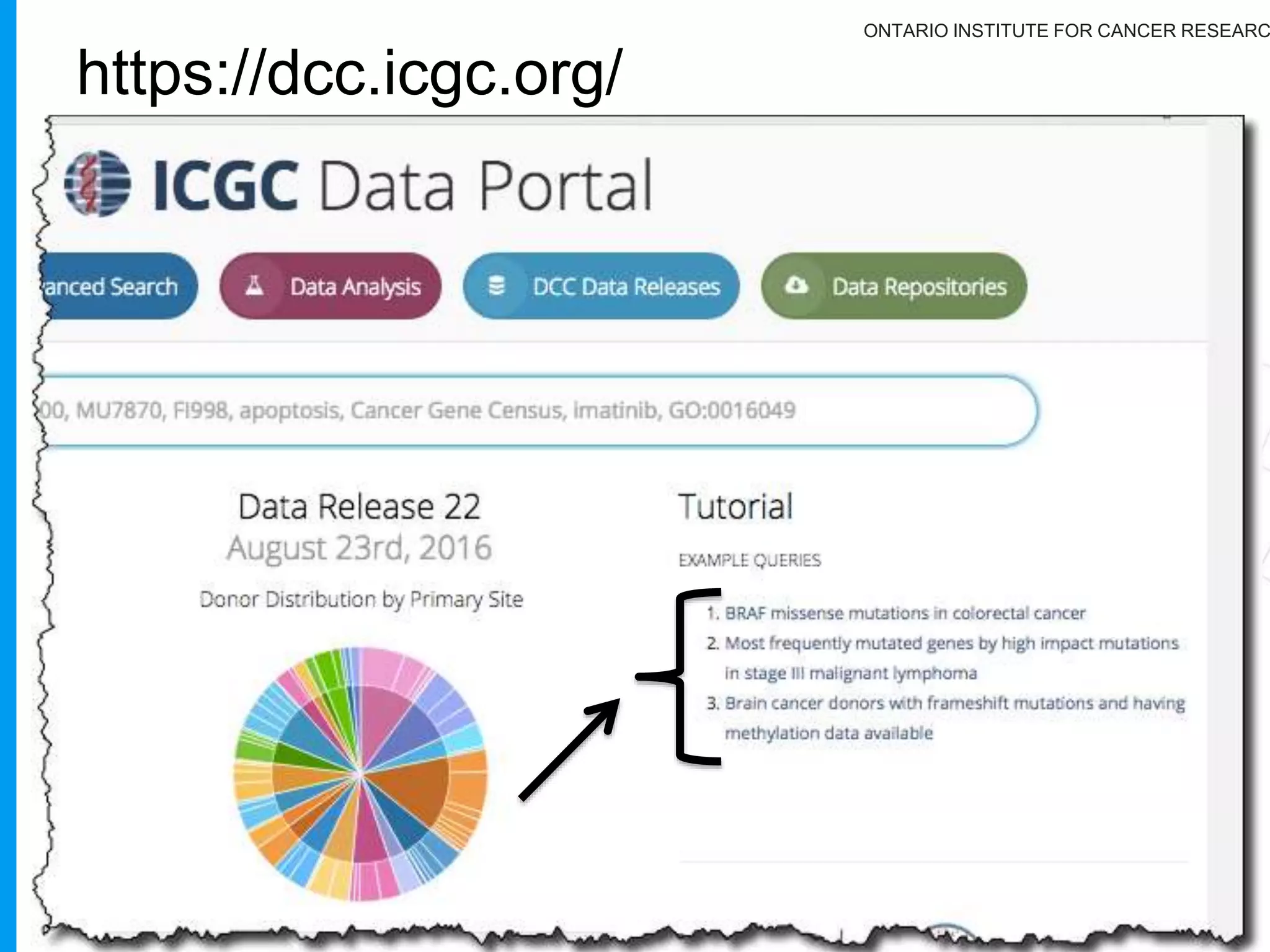
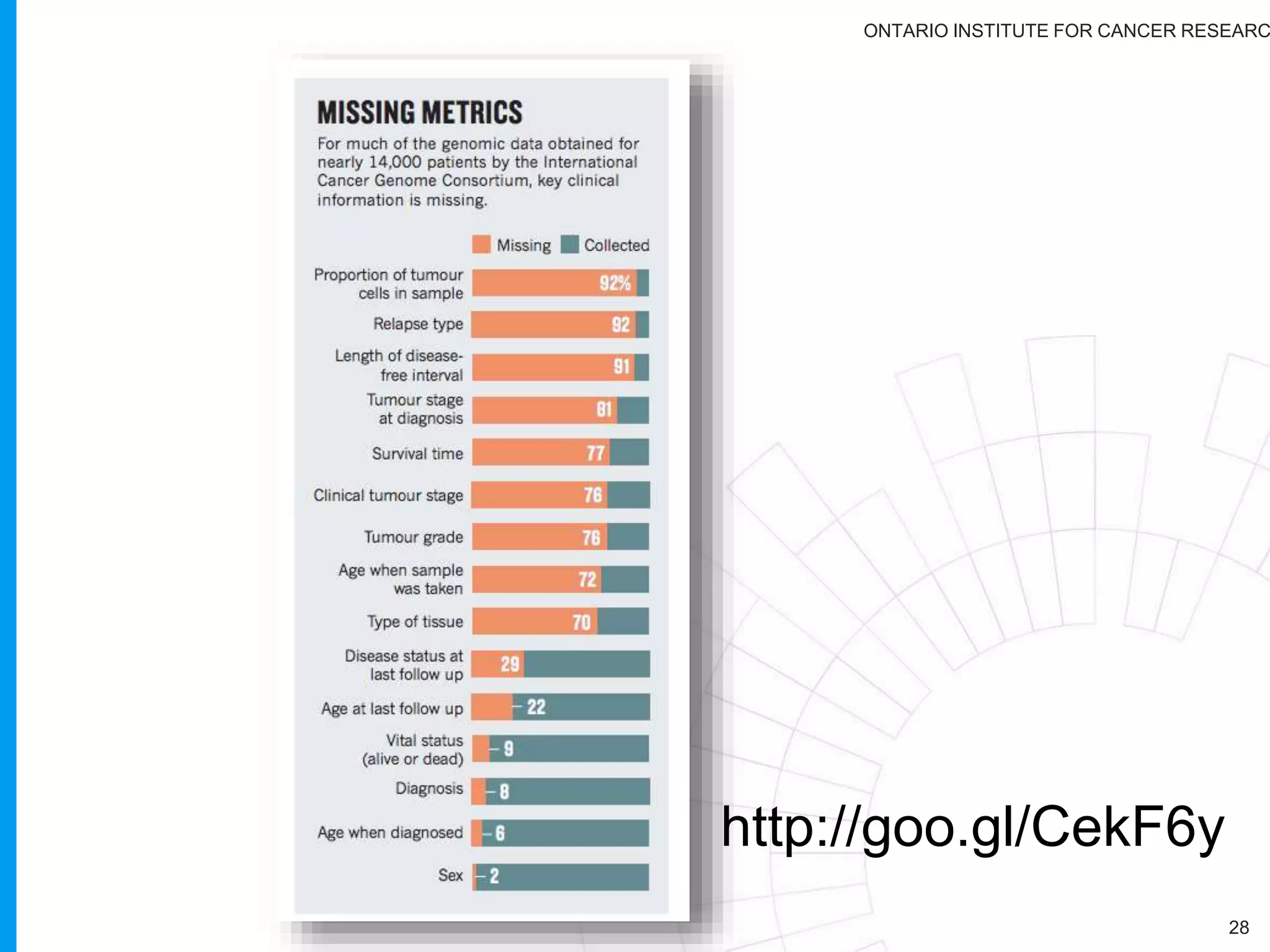
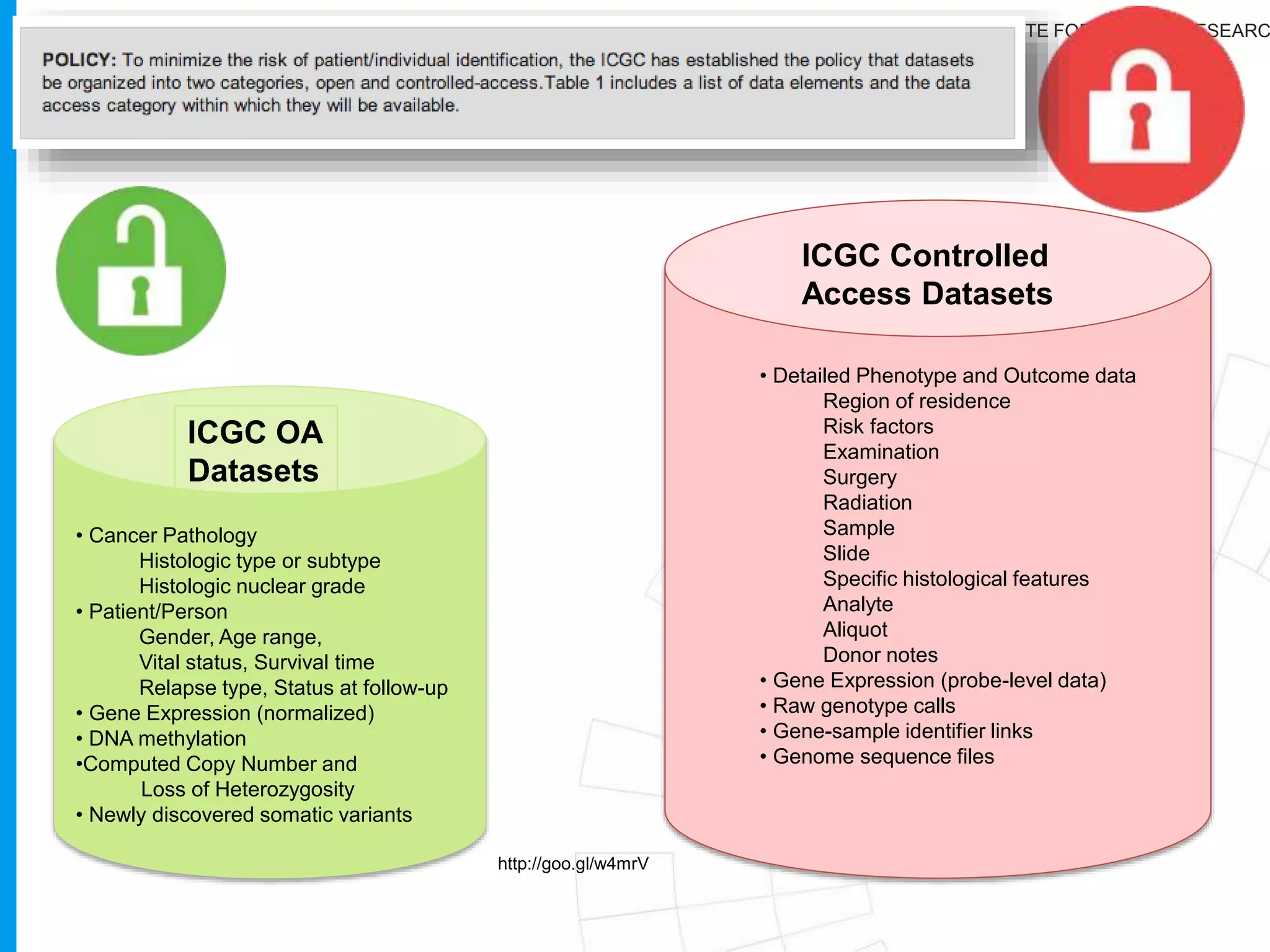
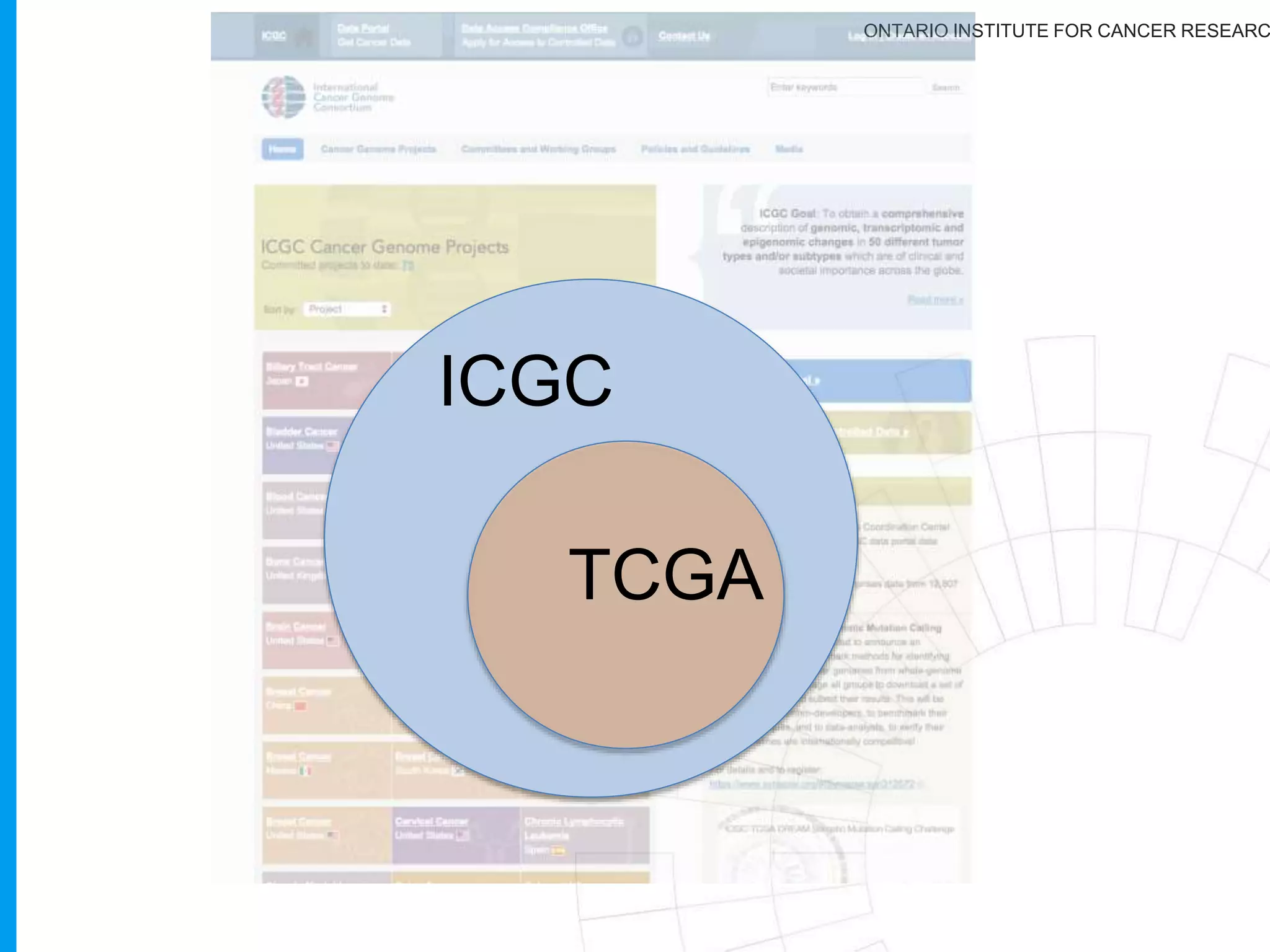
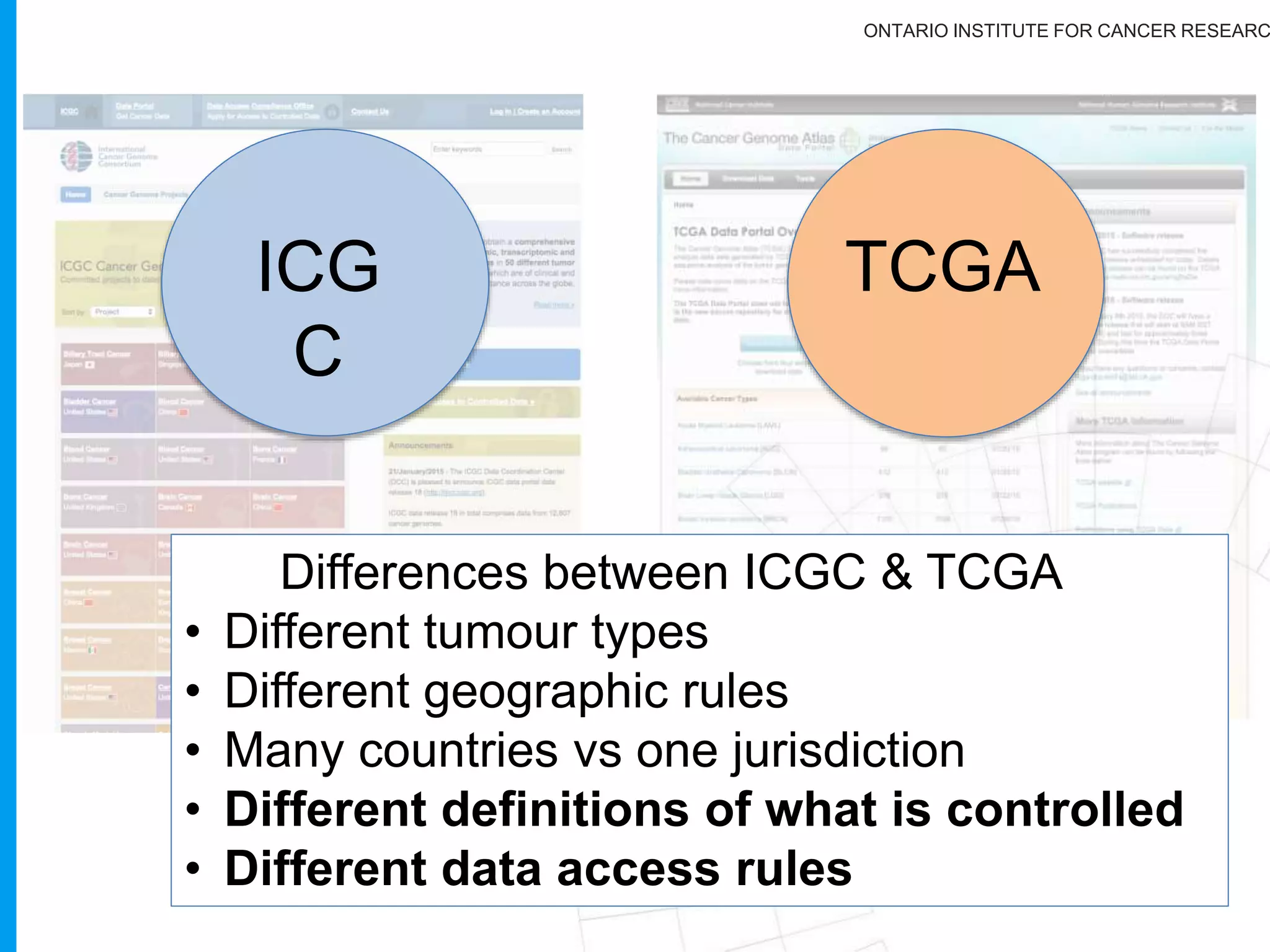
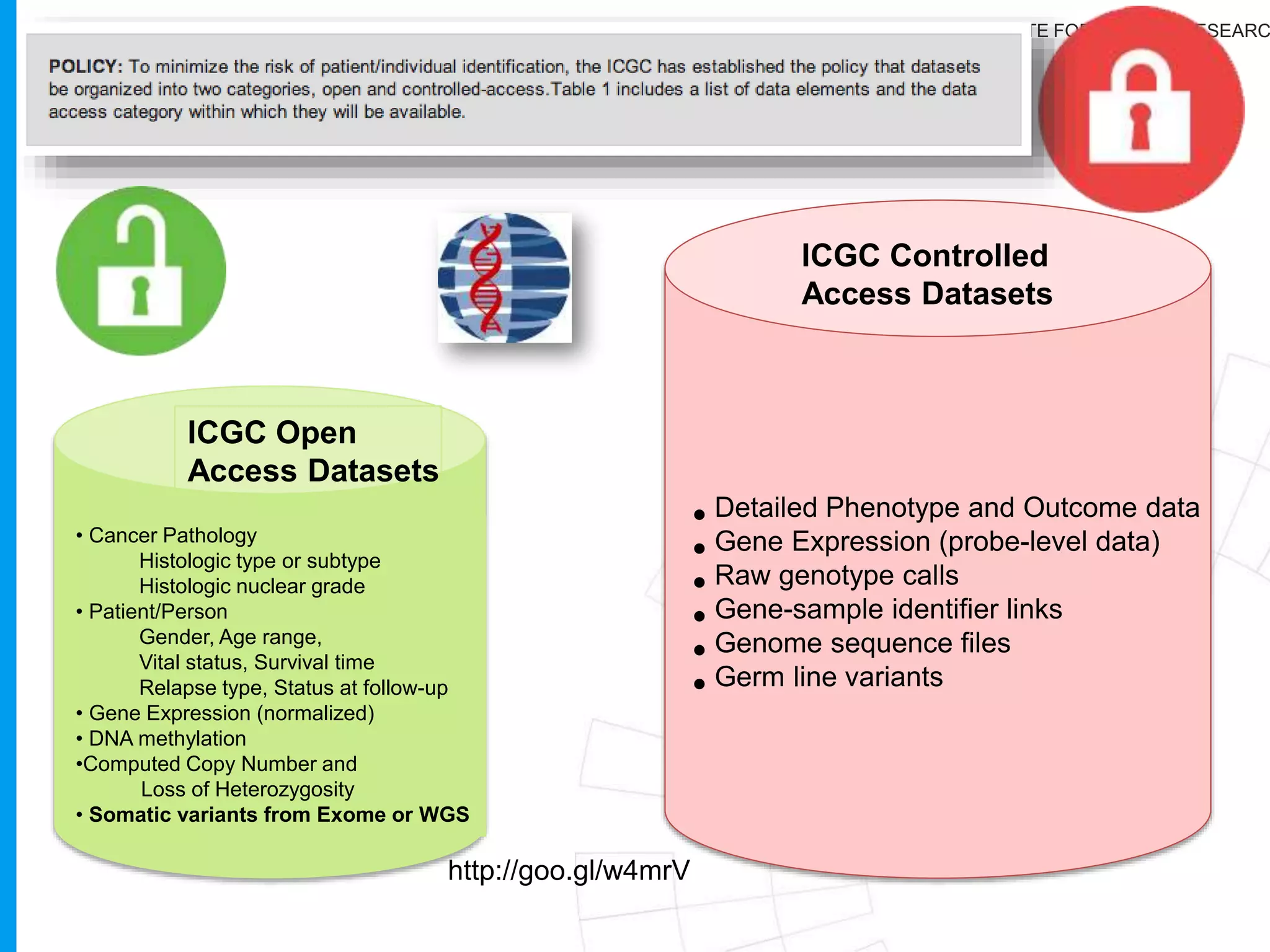
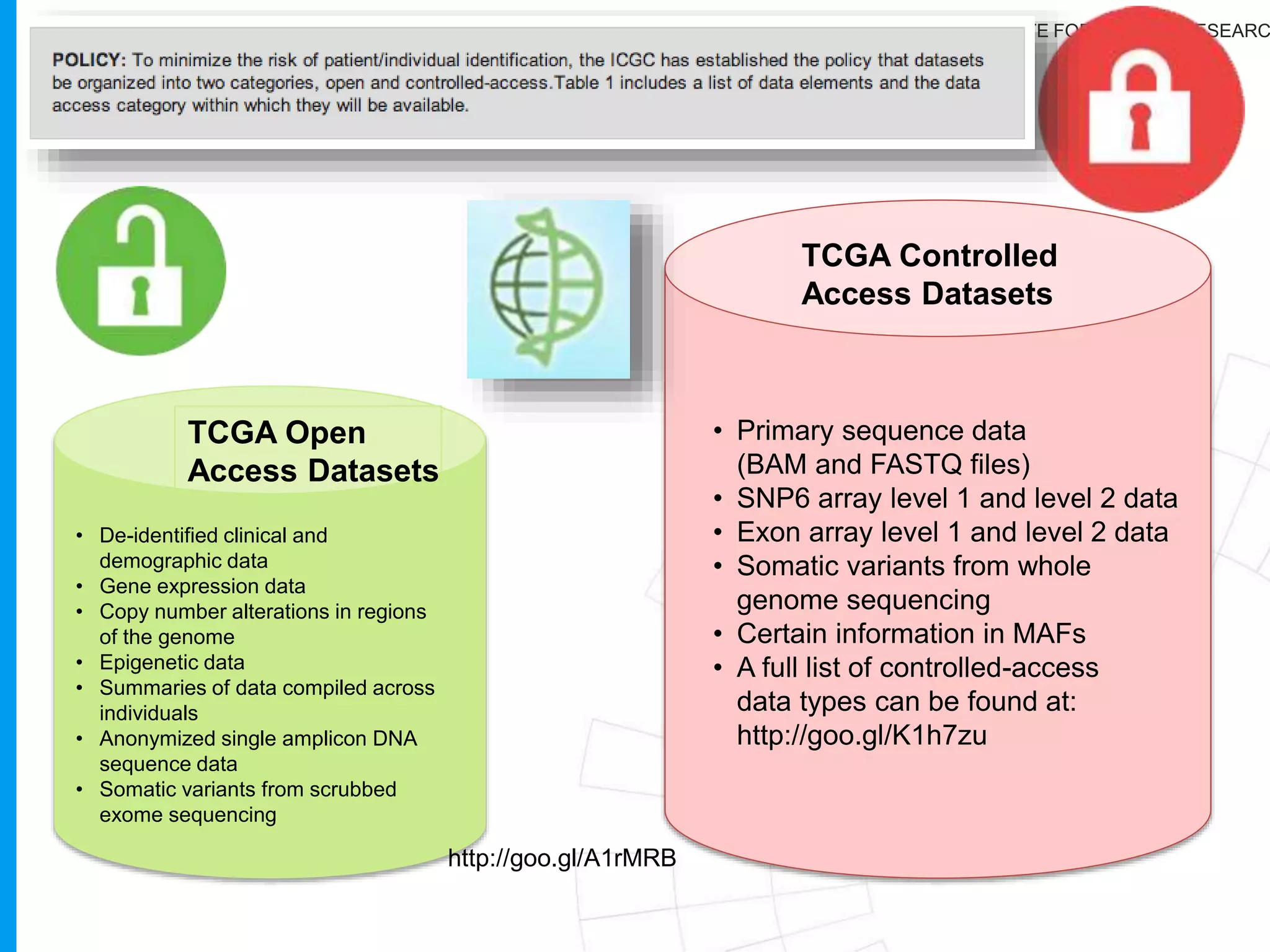
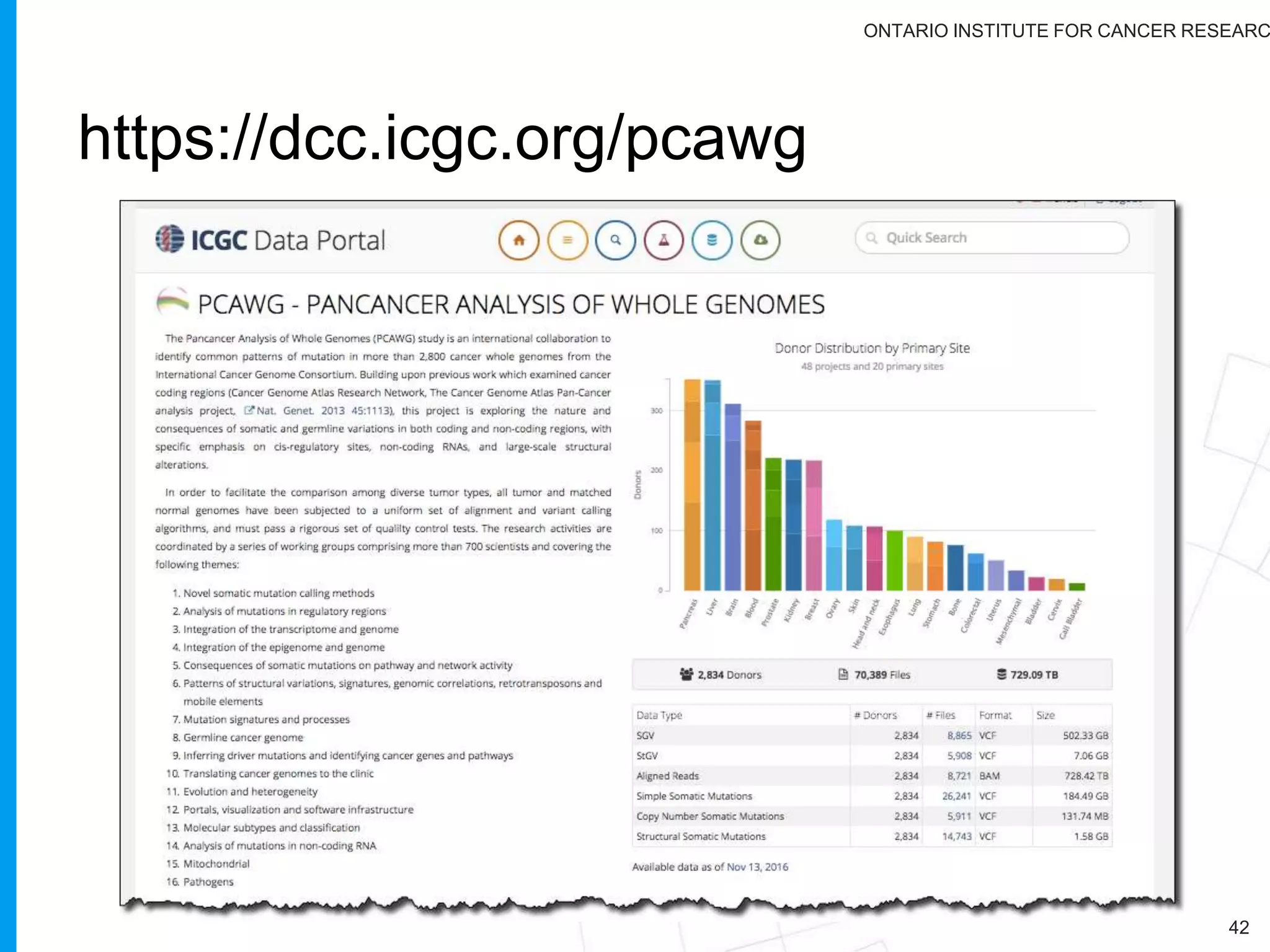
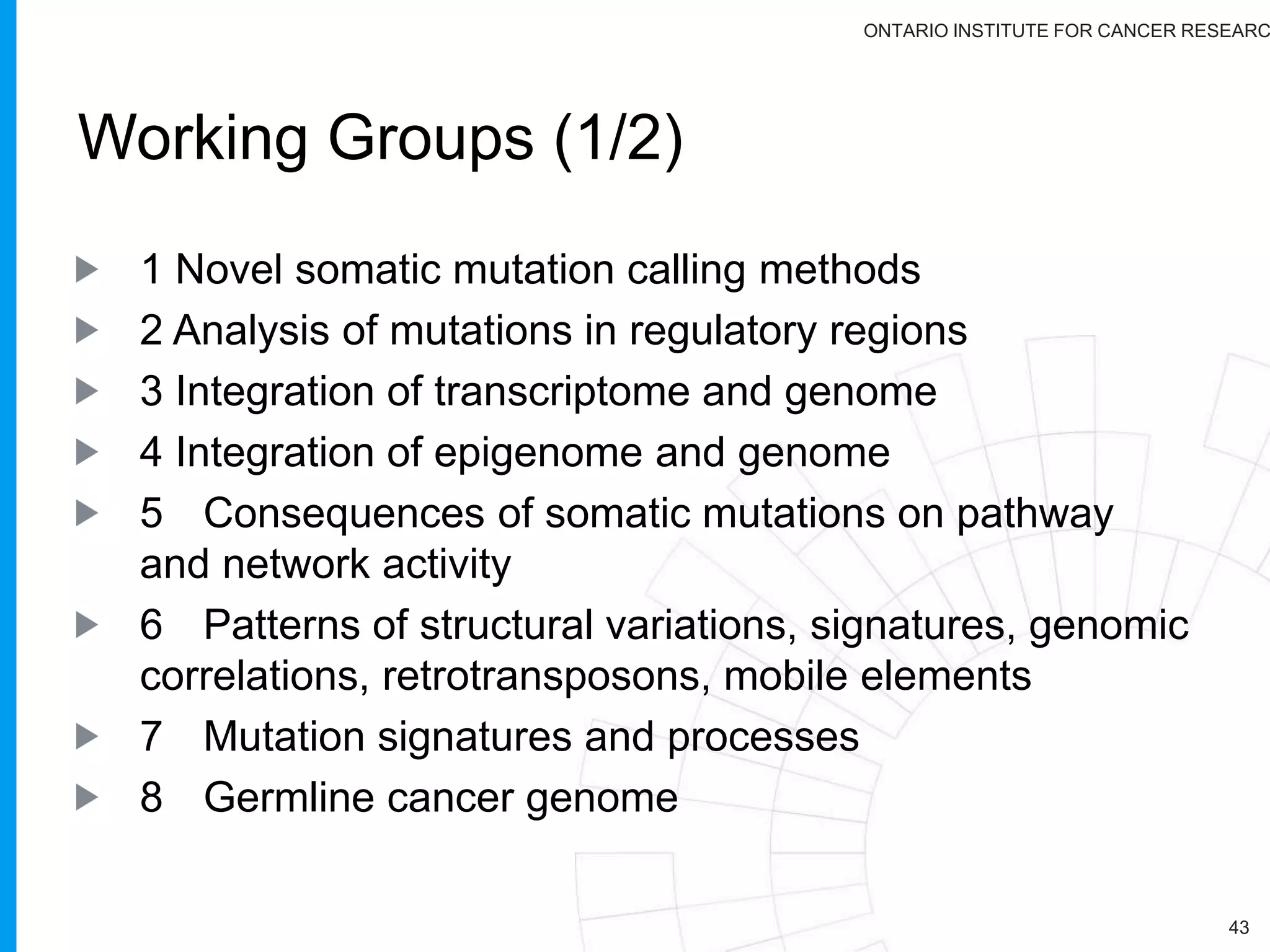
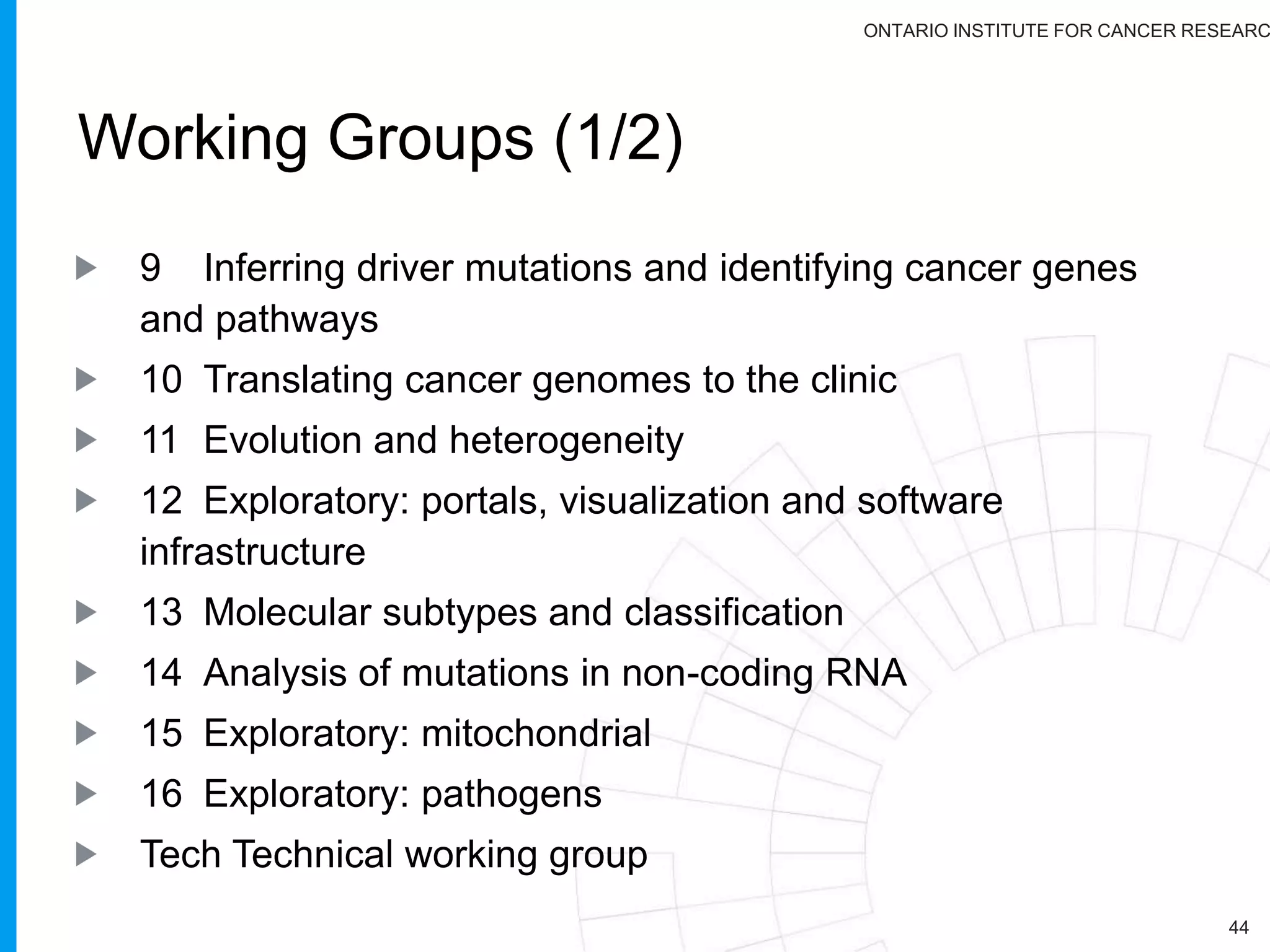
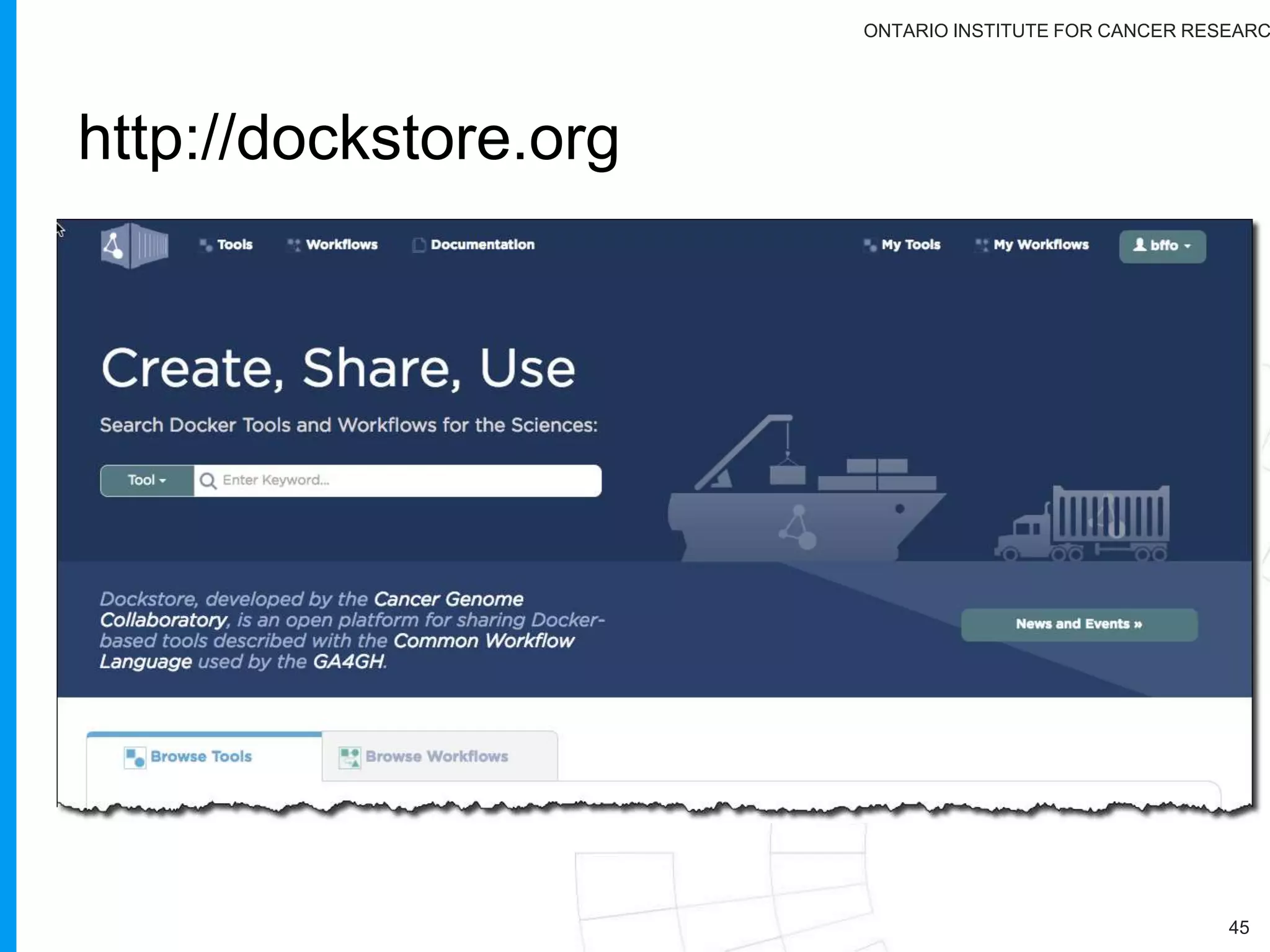
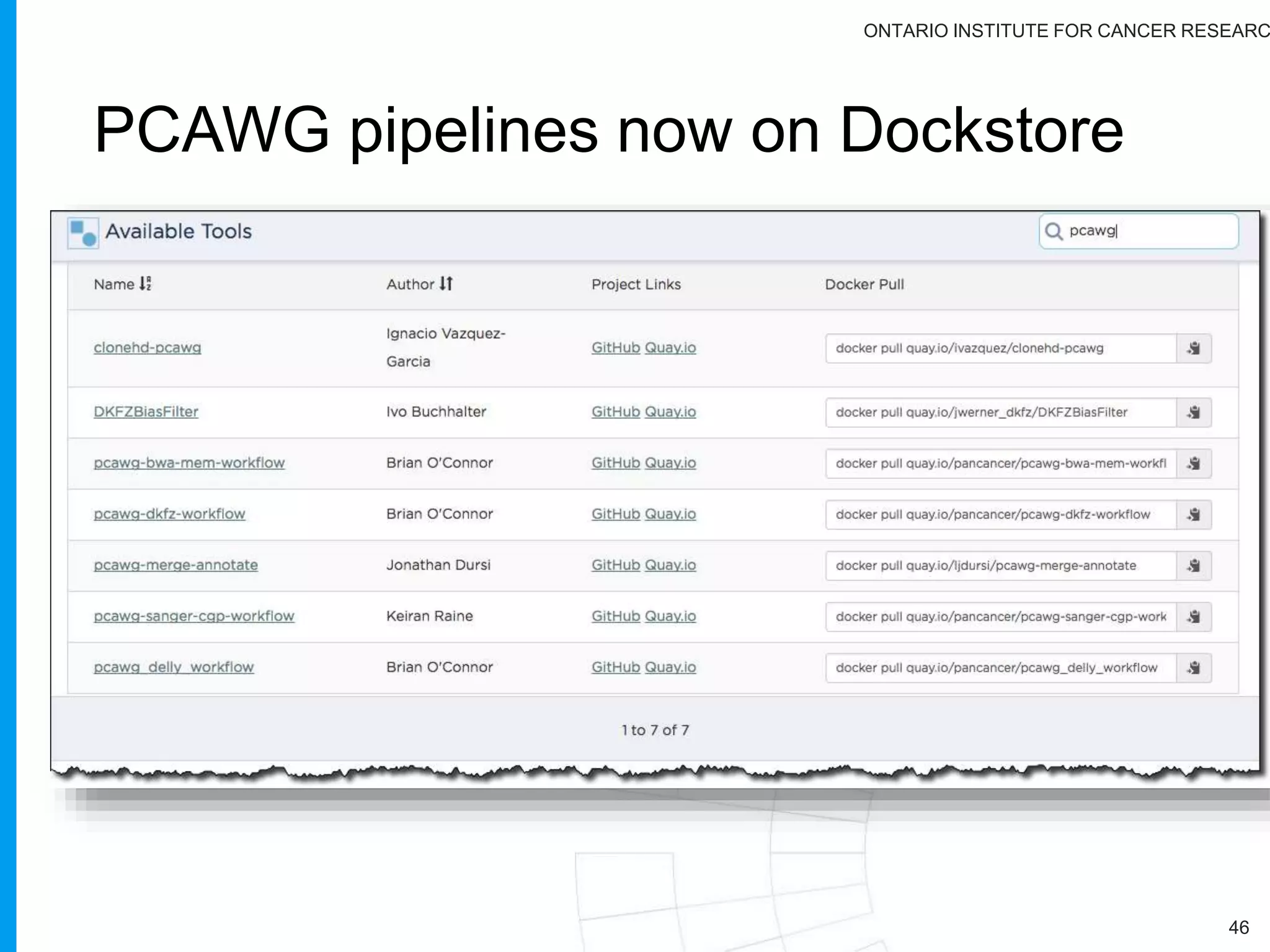
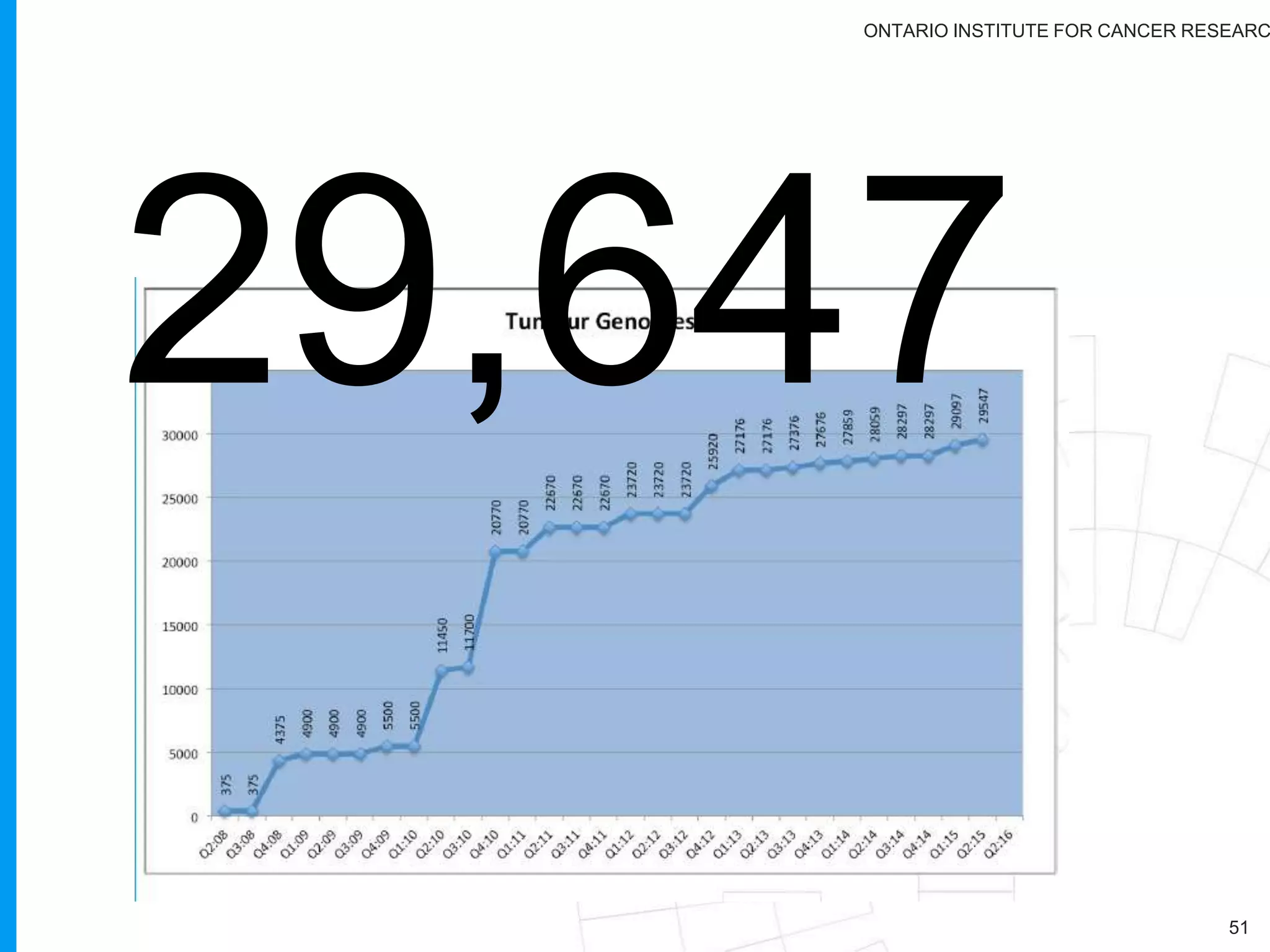
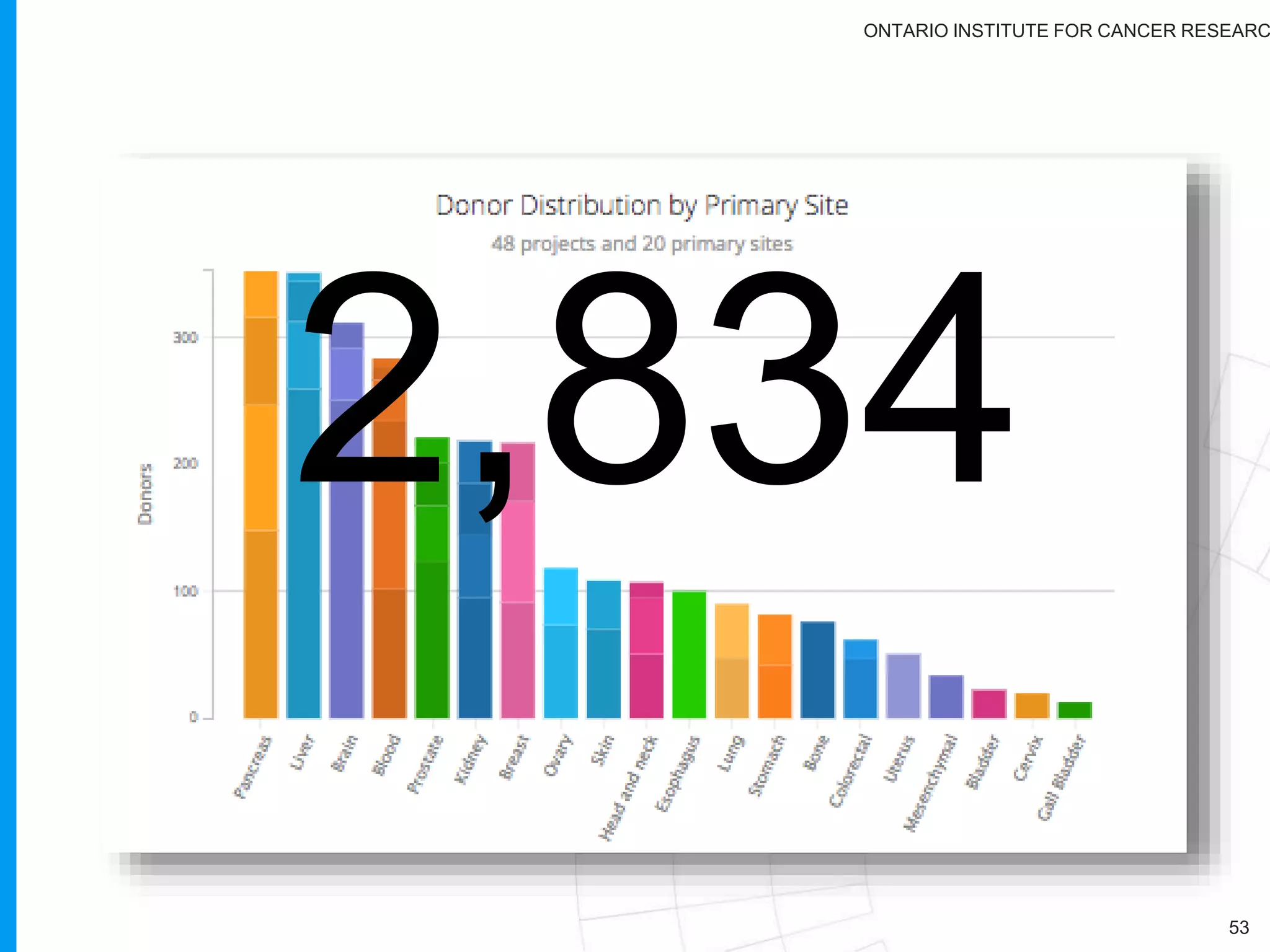
The document provides a report on the Canceromatic III session focusing on pan-cancer analysis and the evolving landscape of data and tools for reproducible cancer genomics workflows. It discusses the challenges of cancer research, the need for standardized methodologies, and the collaborative efforts of the International Cancer Genome Consortium (ICGC) to collect and analyze genomic data across various cancer types. The presentation emphasizes open access to data and the integration of diverse data types to enhance cancer research and treatment strategies.