The document discusses genetic linkage analysis, a statistical method for associating genes with their chromosomal locations. It outlines key concepts like loci, alleles, and inheritance patterns including autosomal and X-linked traits, along with methodologies for conducting linkage mapping. Recent advancements such as the Superlink program are highlighted, which enhances computational efficiency in genetic analysis.




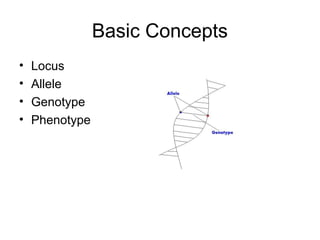







![Main Idea
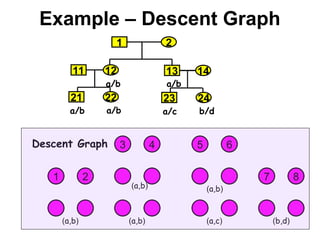
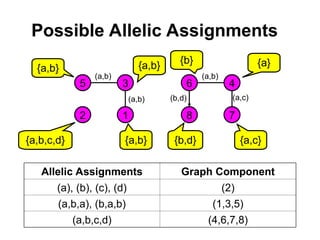
• Let a = (a1,…,a2f) be a vector of alleles assigned to
founders of the pedigree (f is the number of founders).
• We want a graph representation of the restrictions
imposed by the observed marker genotypes on the
vector a that can be assigned to the founder genes.
• The algorithm extracts only vectors a compatible with
the marker data.
• Pr[m|v] is obtained via a sum over all compatible vectors
a.](https://image.slidesharecdn.com/linkageanalysis-240909051202-8b5f6b0b/85/LinkageAnalysis-pptmjbhjxfzfxghvjbknknklm-22-320.jpg)
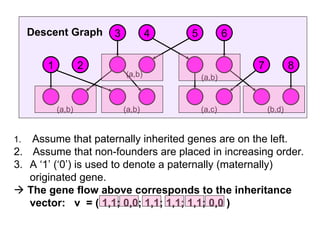
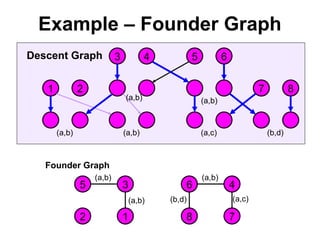
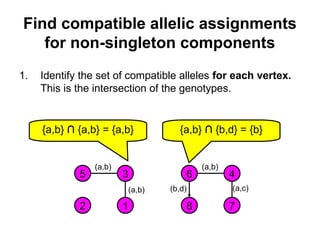
![Computing P(m|v)
• If for some component there are no possible allelic
assignments, then P(m|v) = 0.
• The probability of singleton components is 1 we can
ignore them.
• Let ahi be an element of a vector of alleles assigned to the
vertices of component Ci.
]
Pr[
]
Pr[
}
:
{
i
C
j
j
j
hi a
a
]
Pr[
]
Pr[
}
:
{
i
hi A
a
h
hi
i a
C
]
Pr[
]
|
Pr[
1
m
i
i
C
v
m
over 2f elements
2
terms at most
Linear in the number of founders](https://image.slidesharecdn.com/linkageanalysis-240909051202-8b5f6b0b/85/LinkageAnalysis-pptmjbhjxfzfxghvjbknknklm-29-320.jpg)