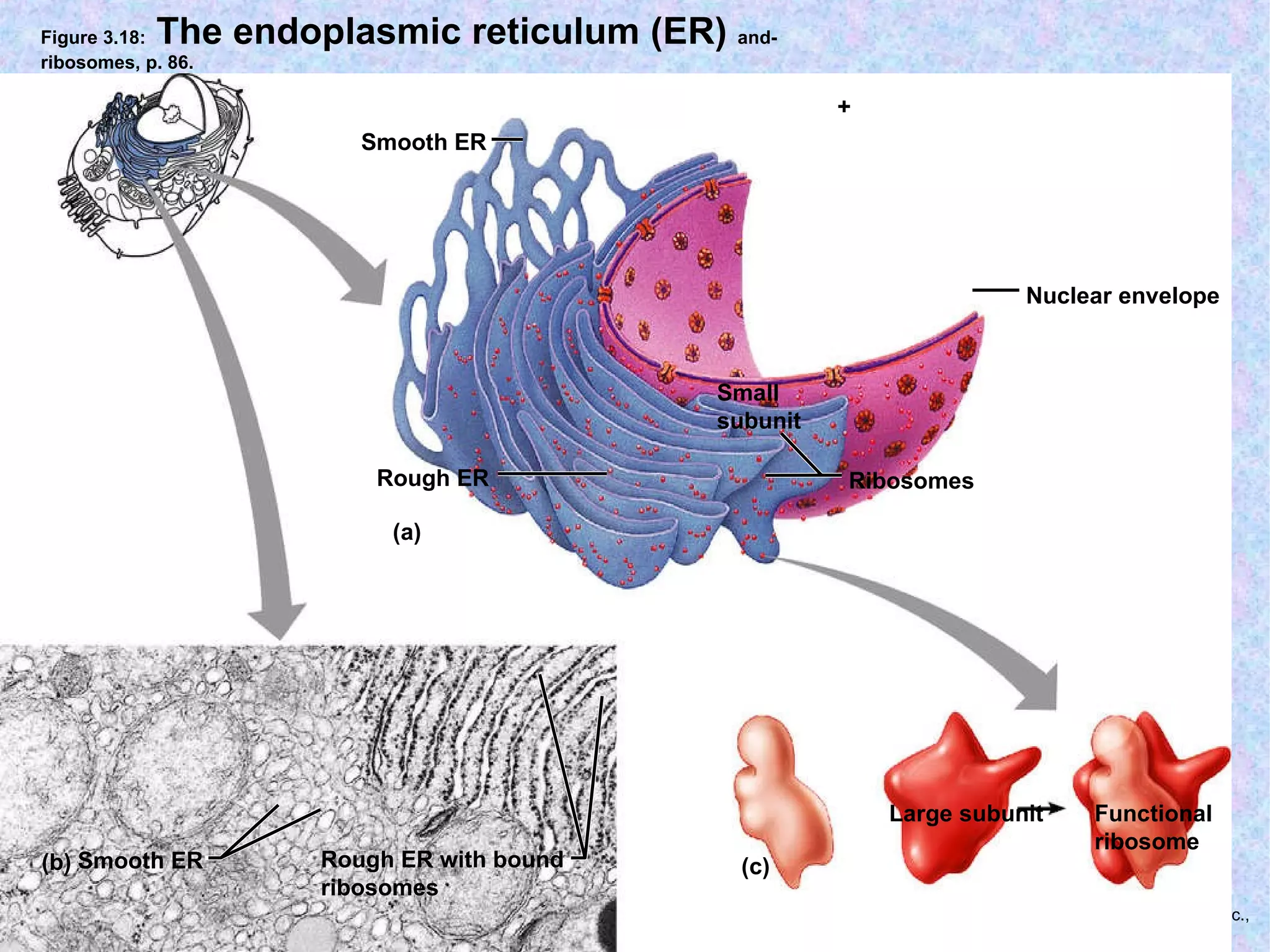
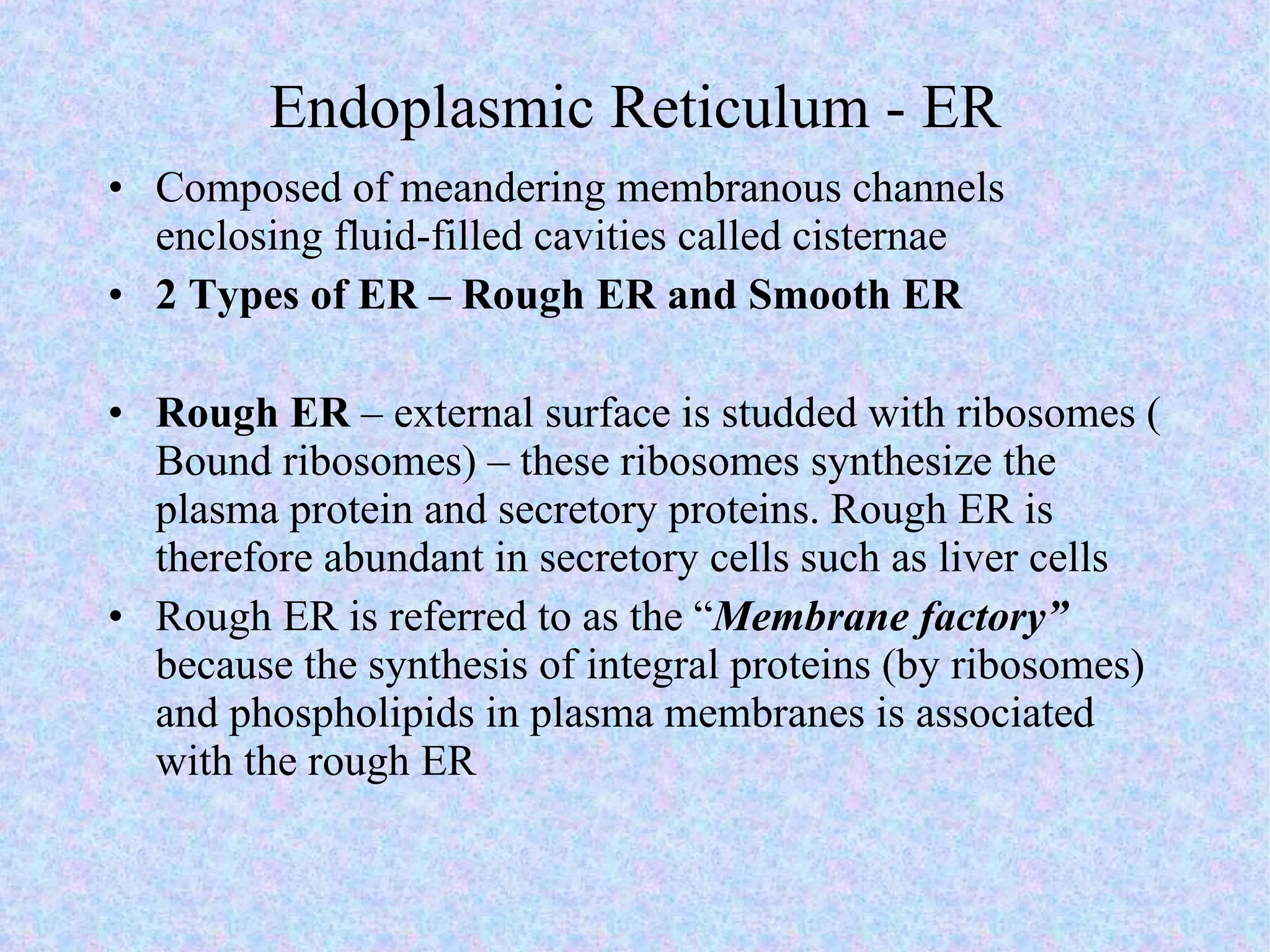
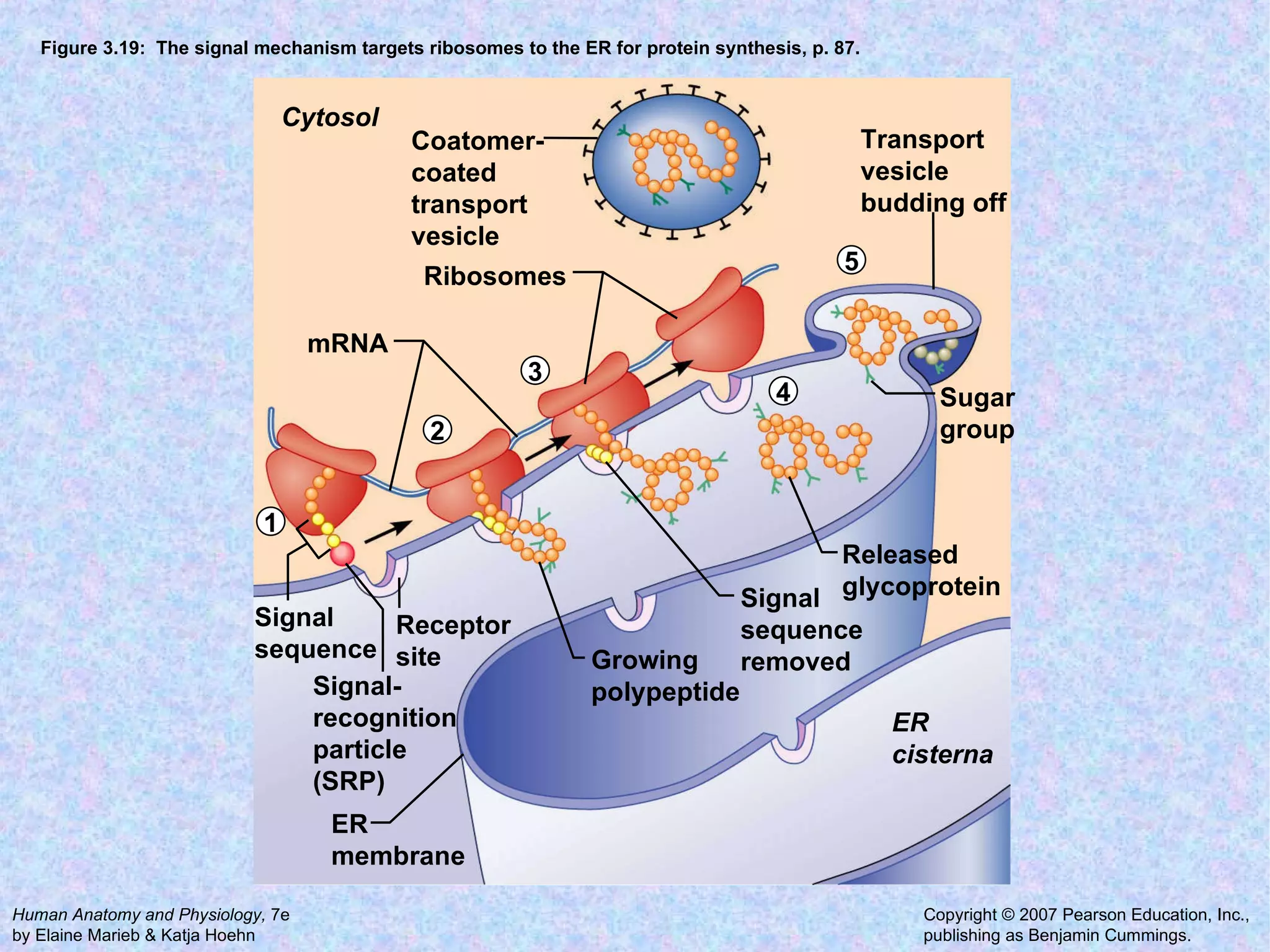
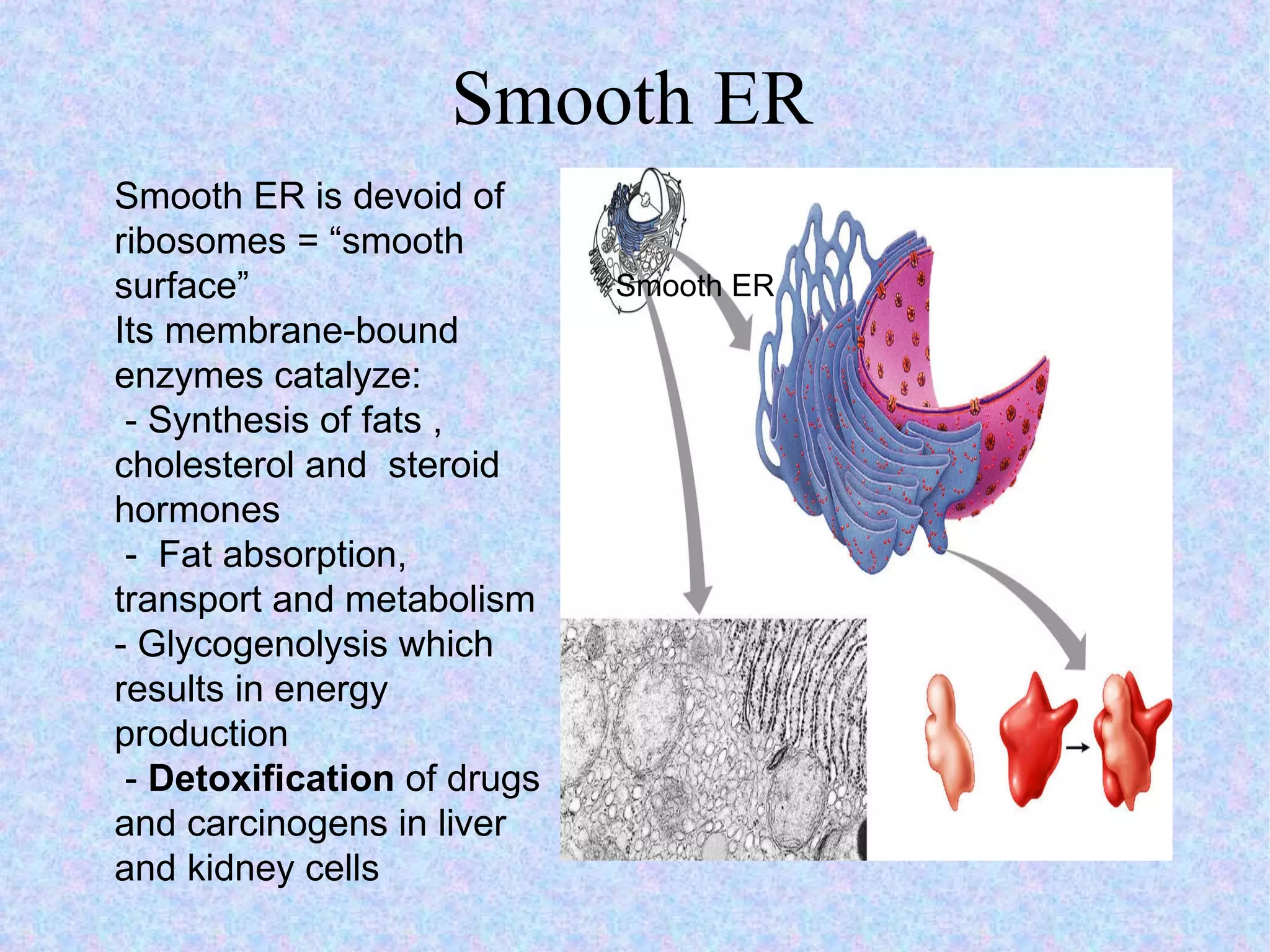
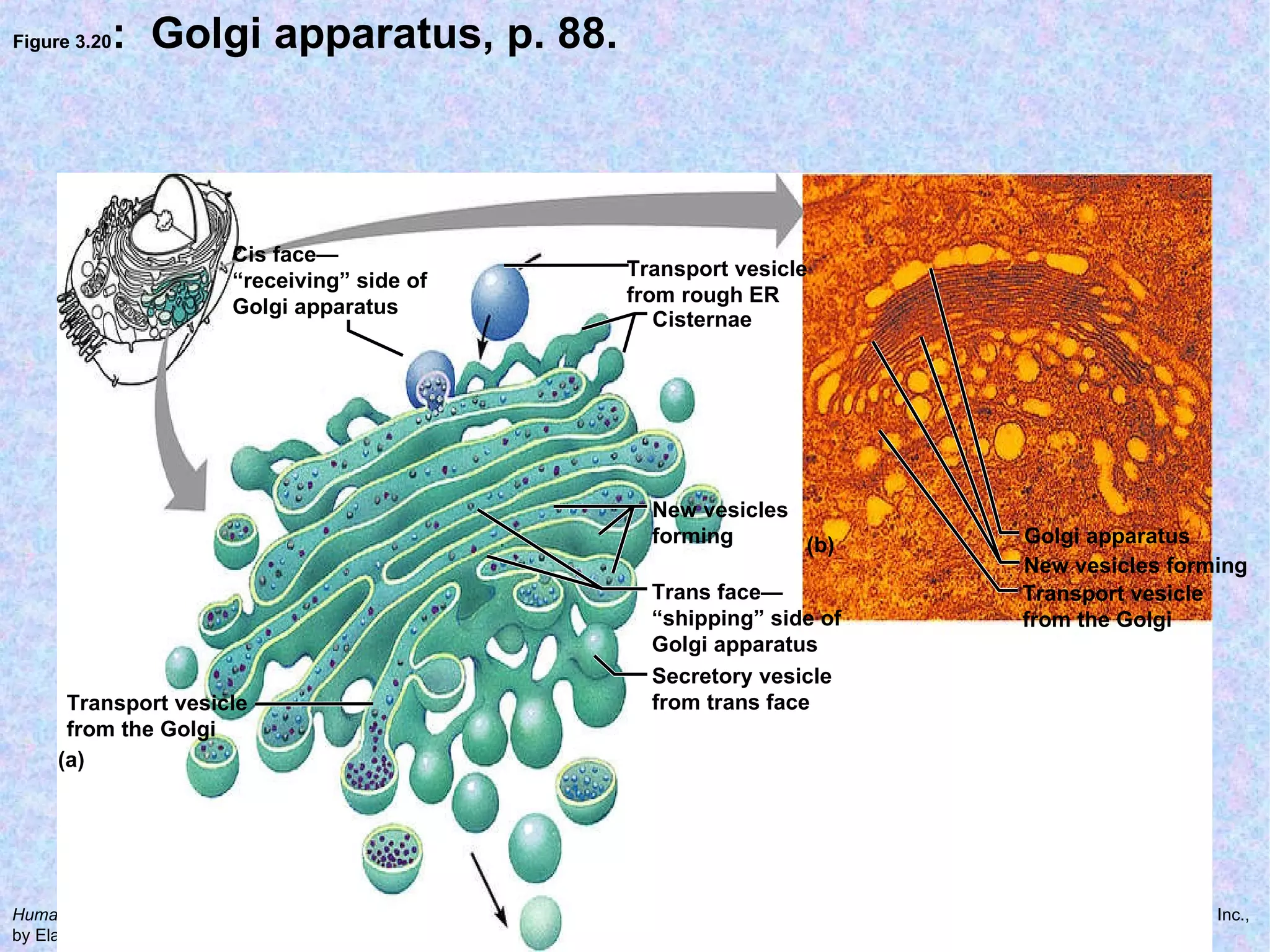
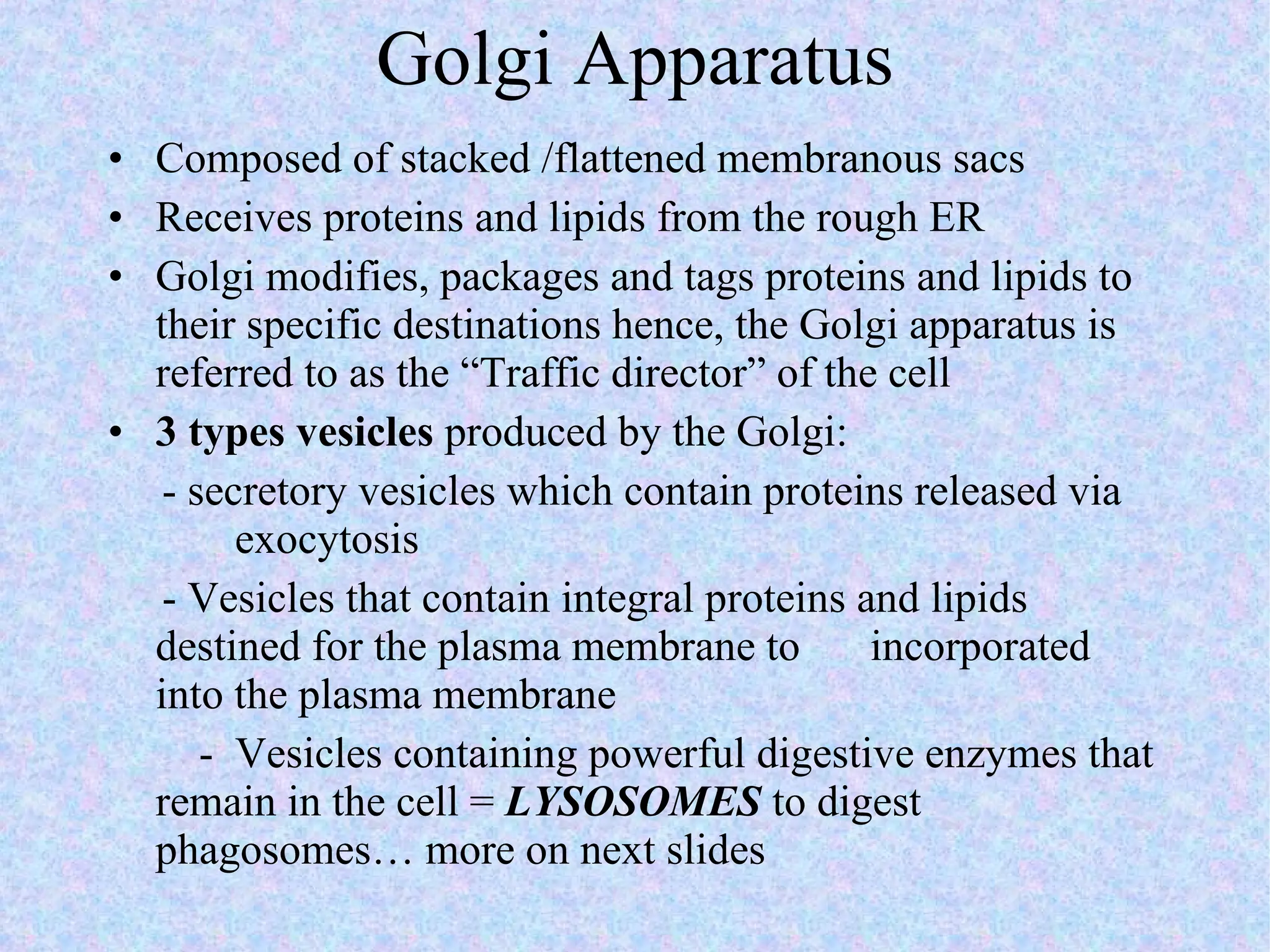
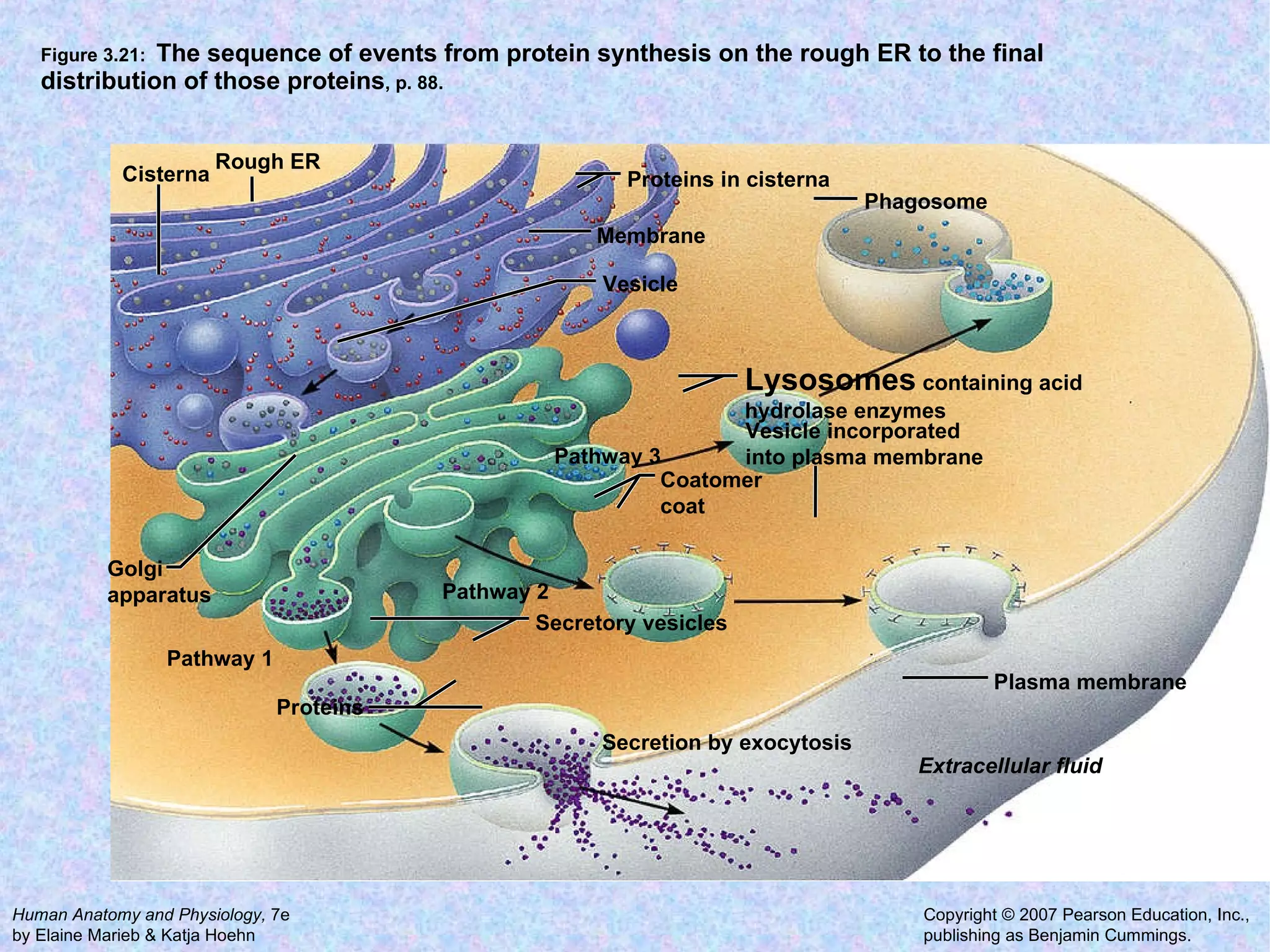
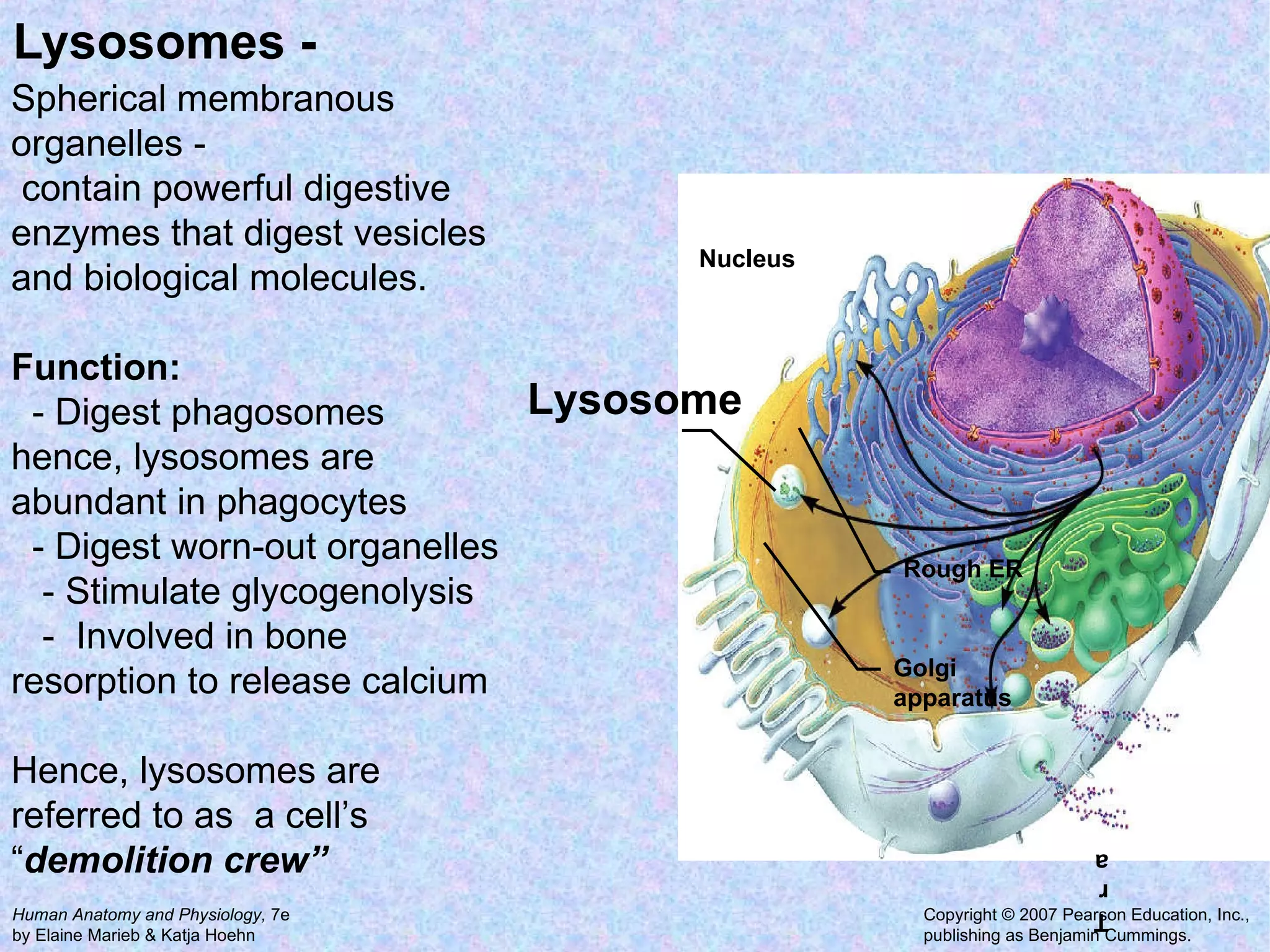
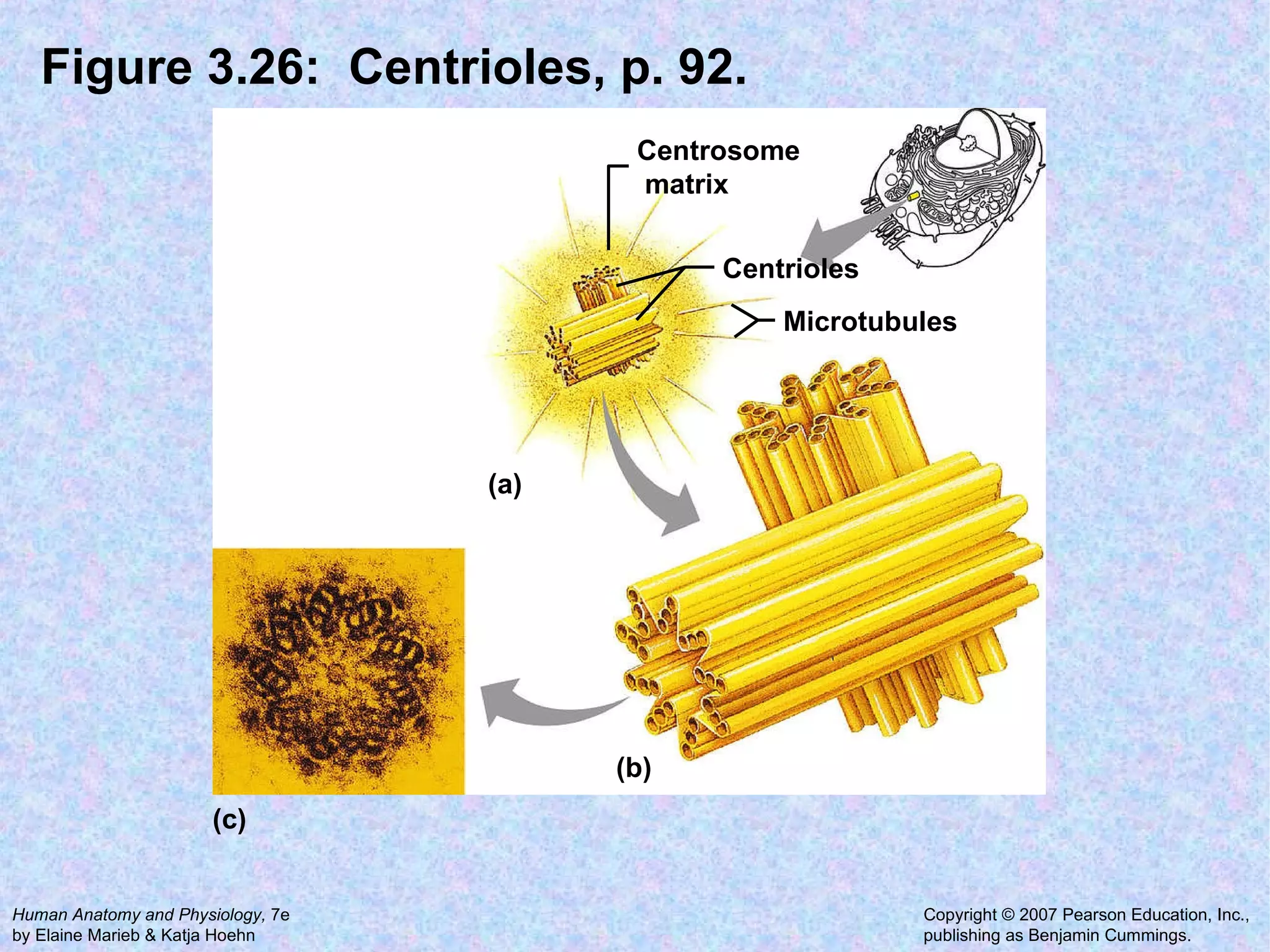
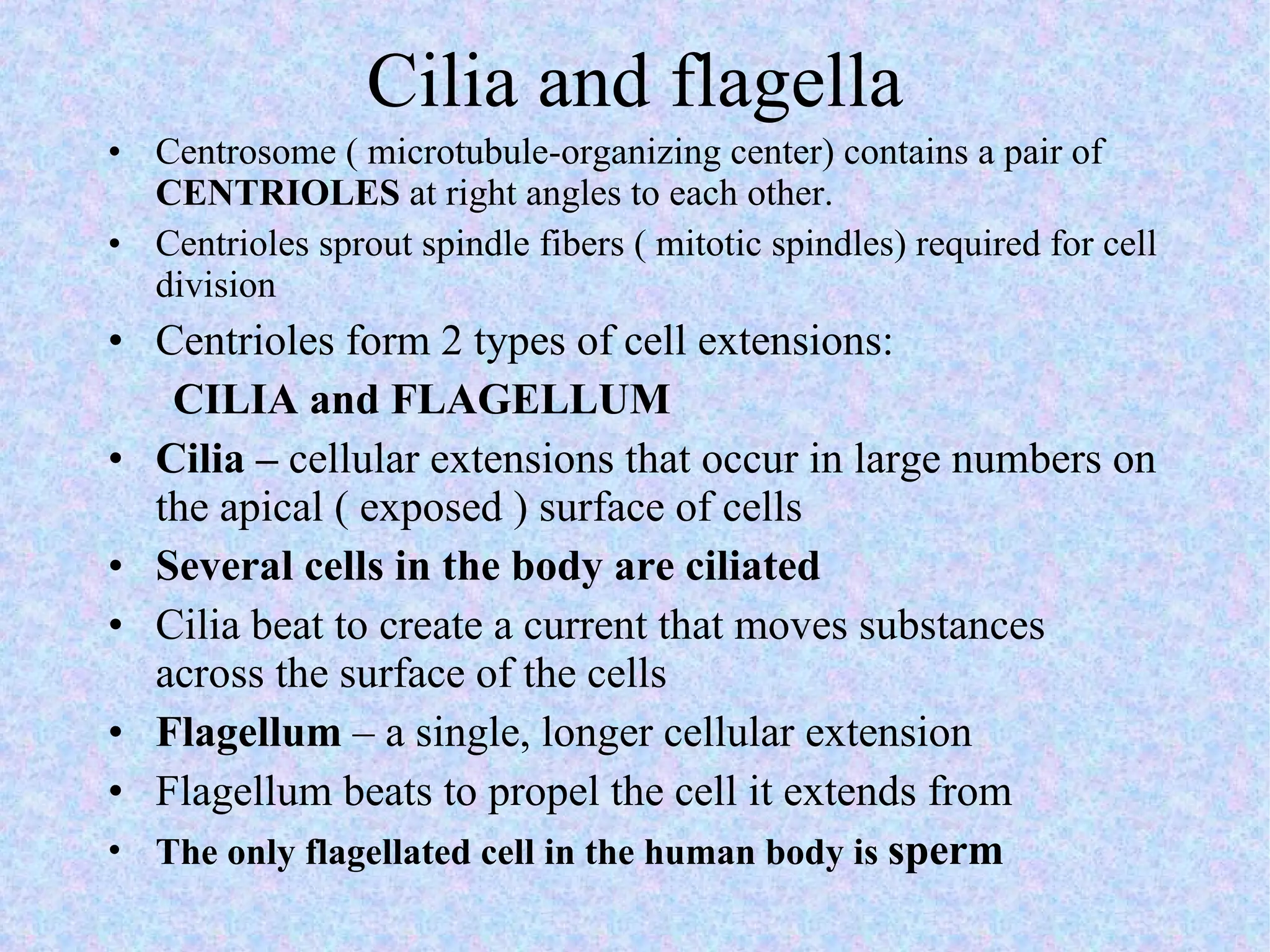
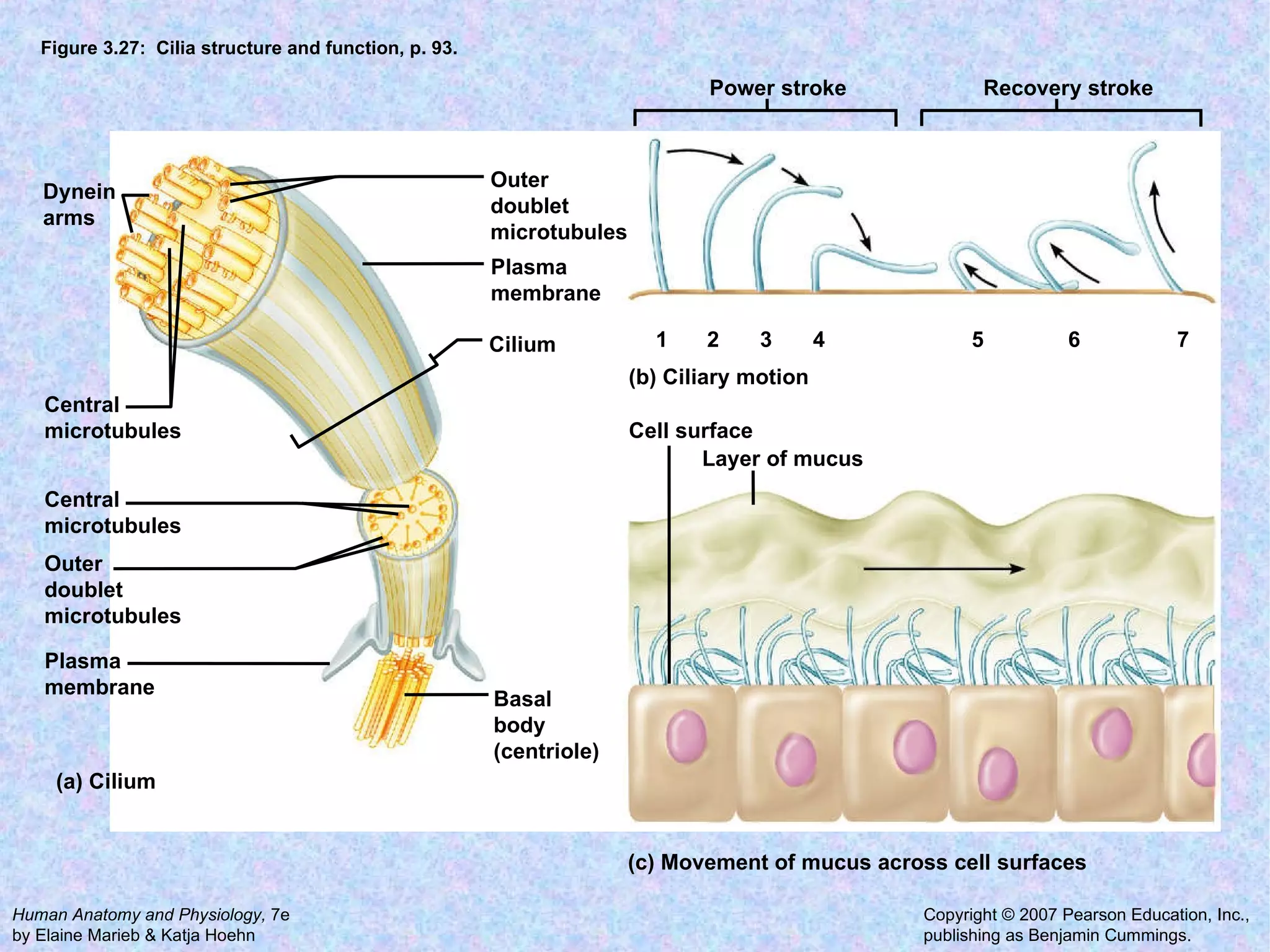
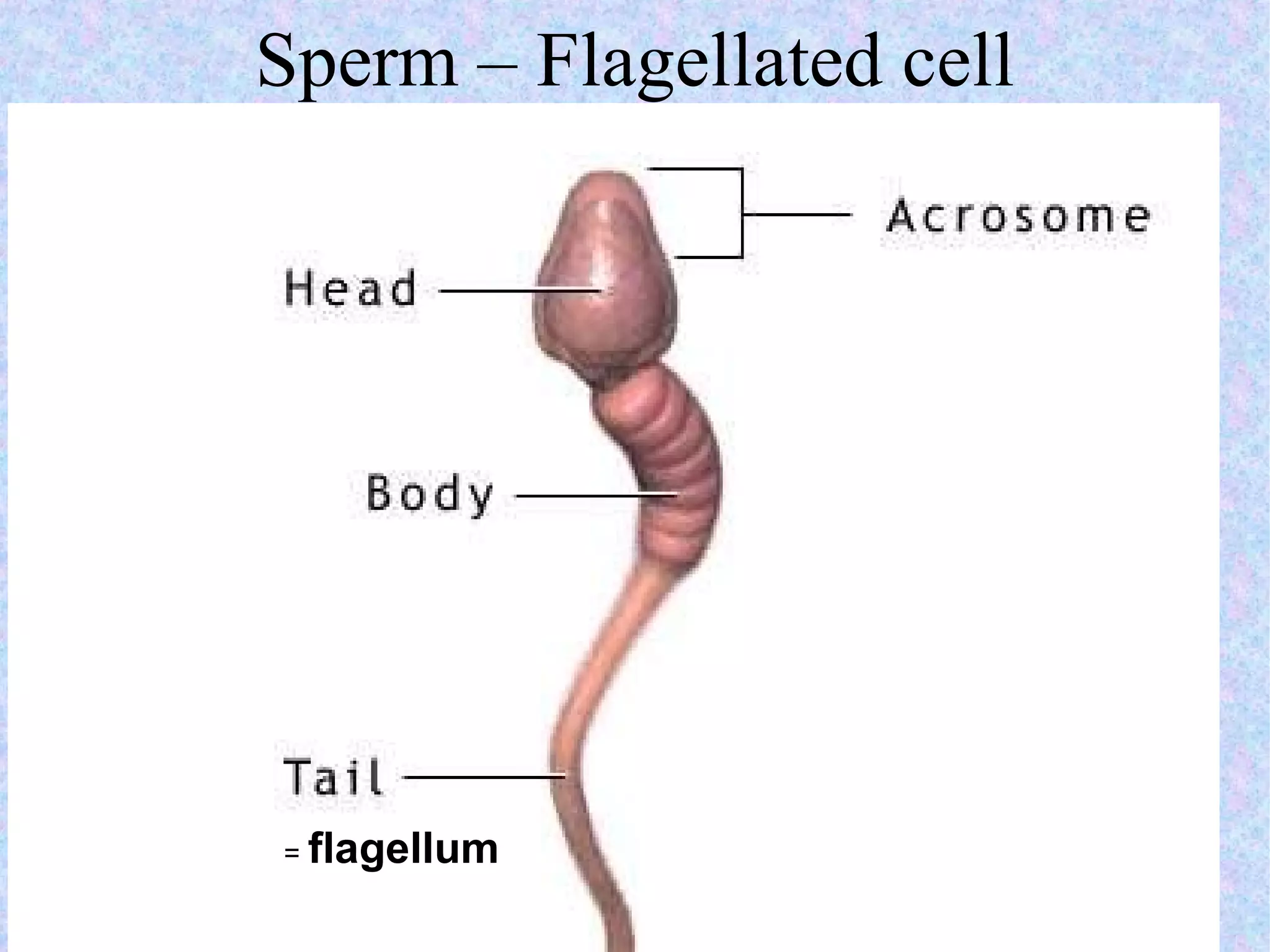
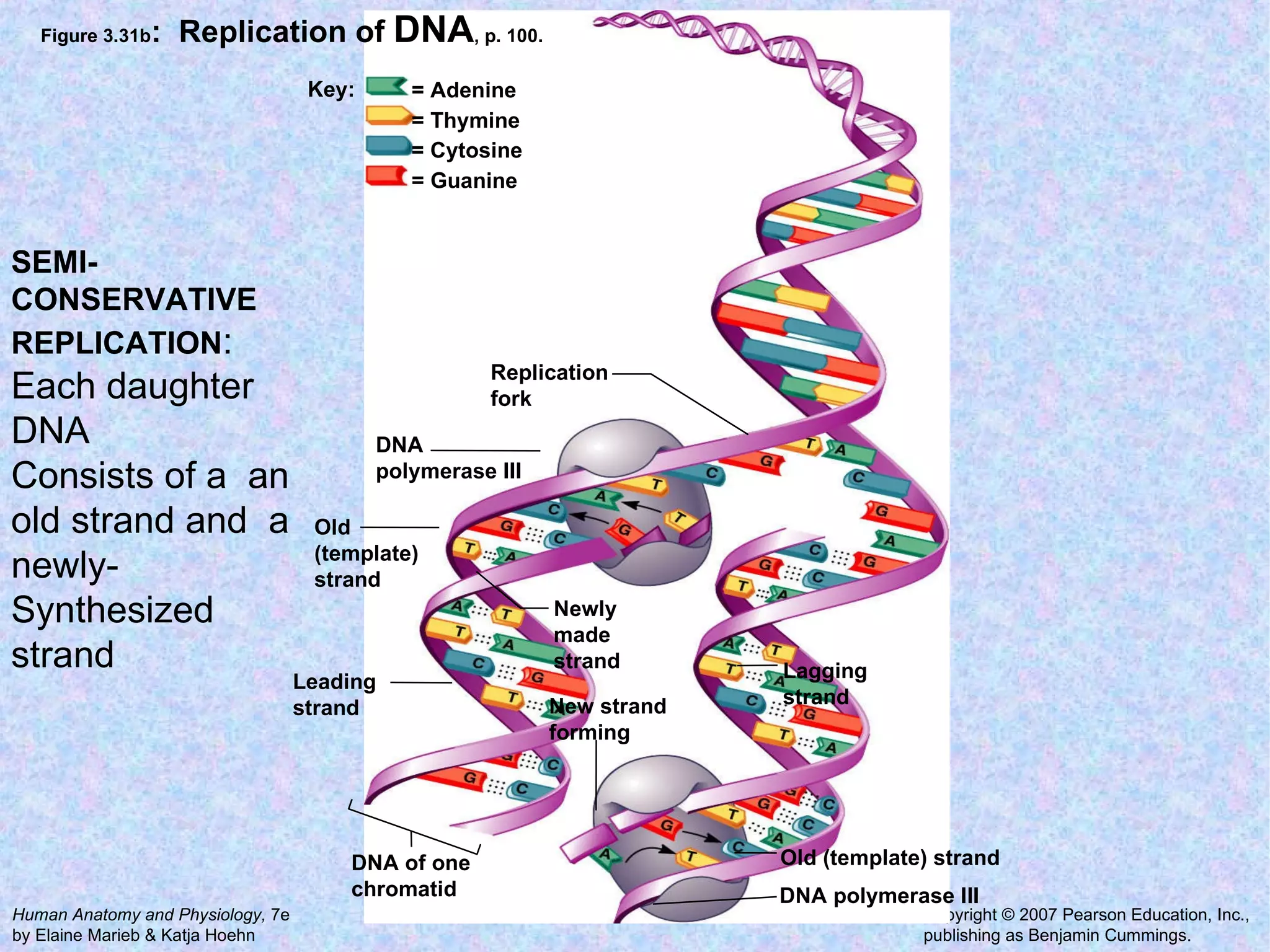
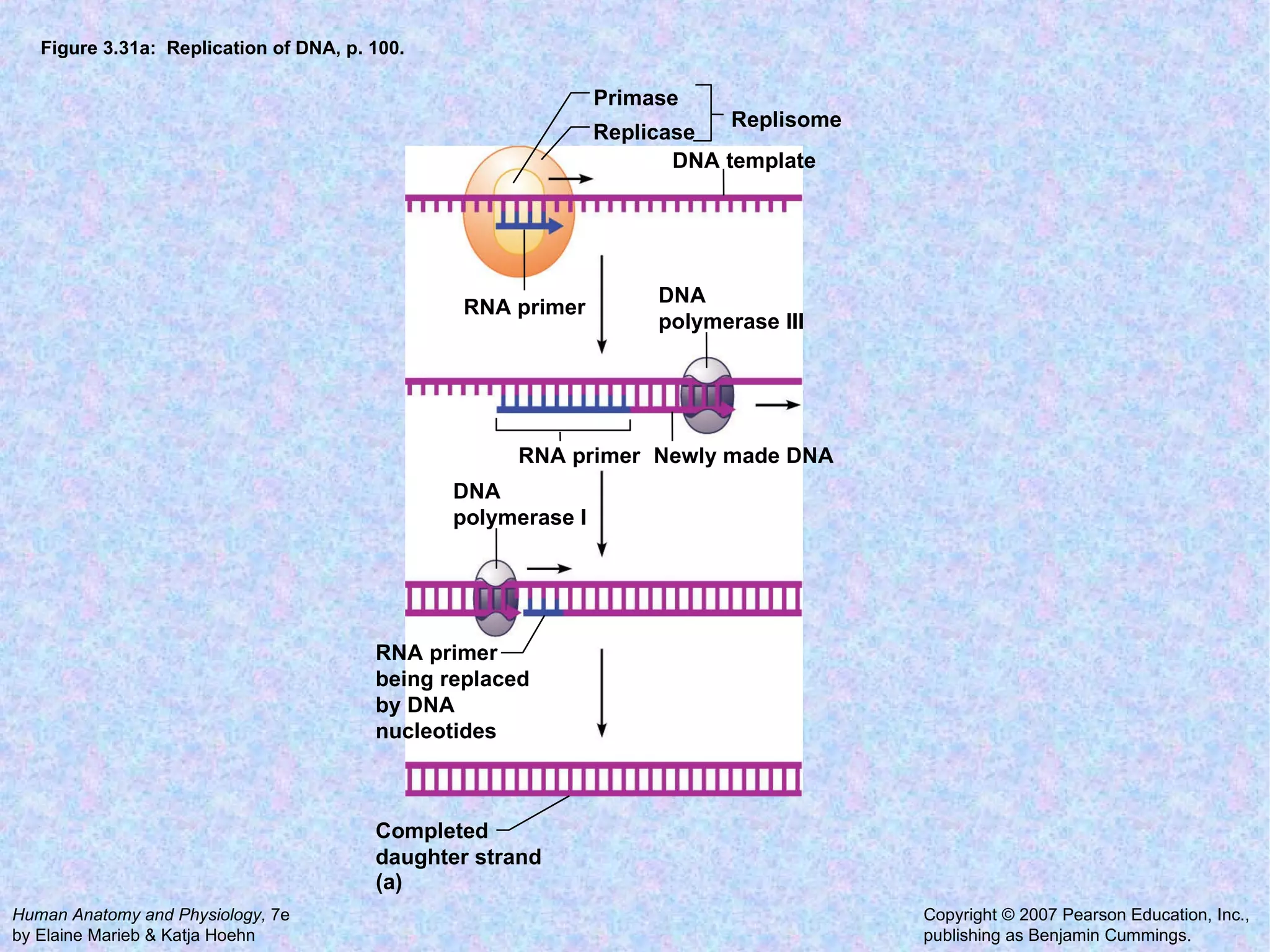
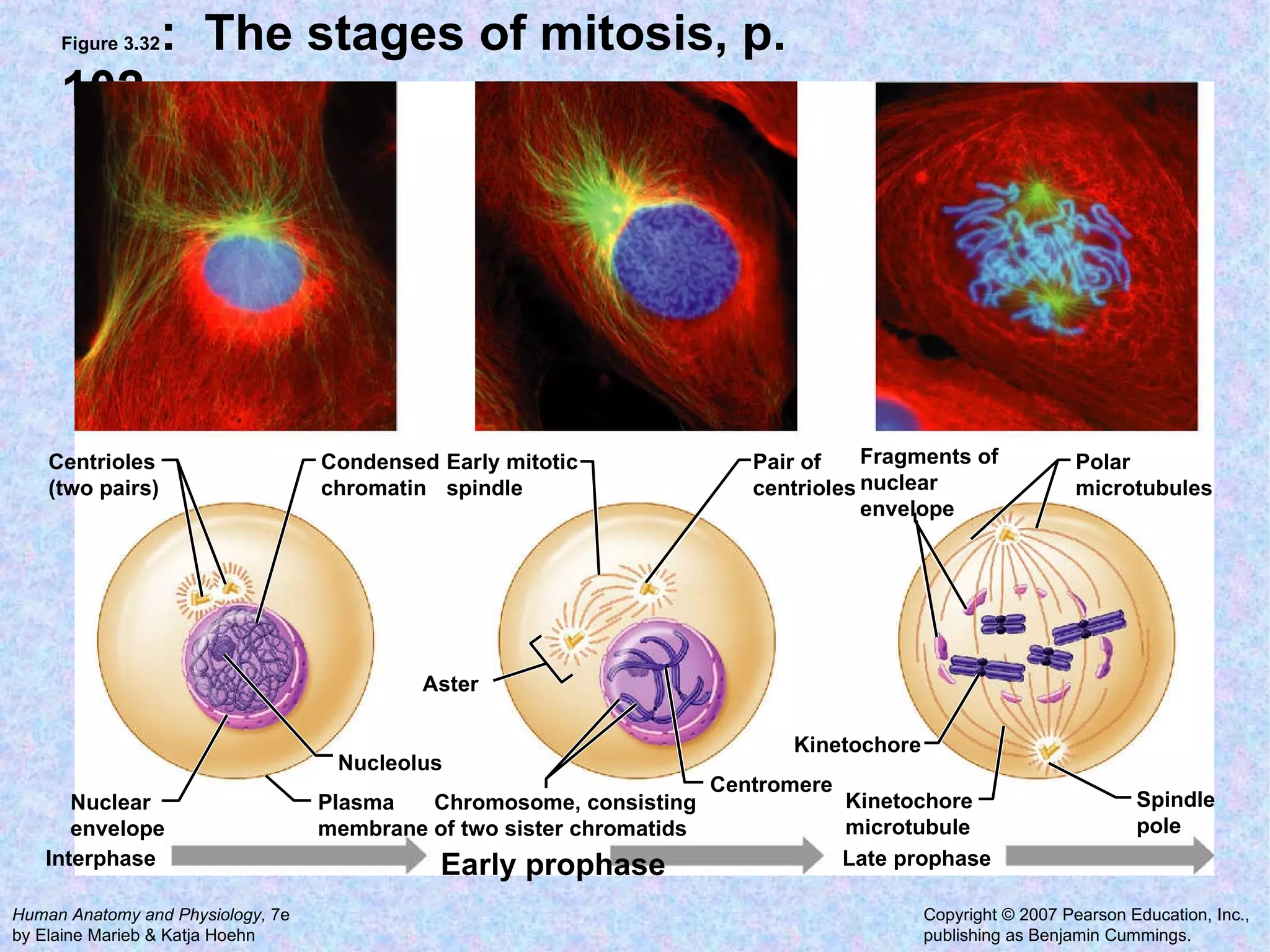
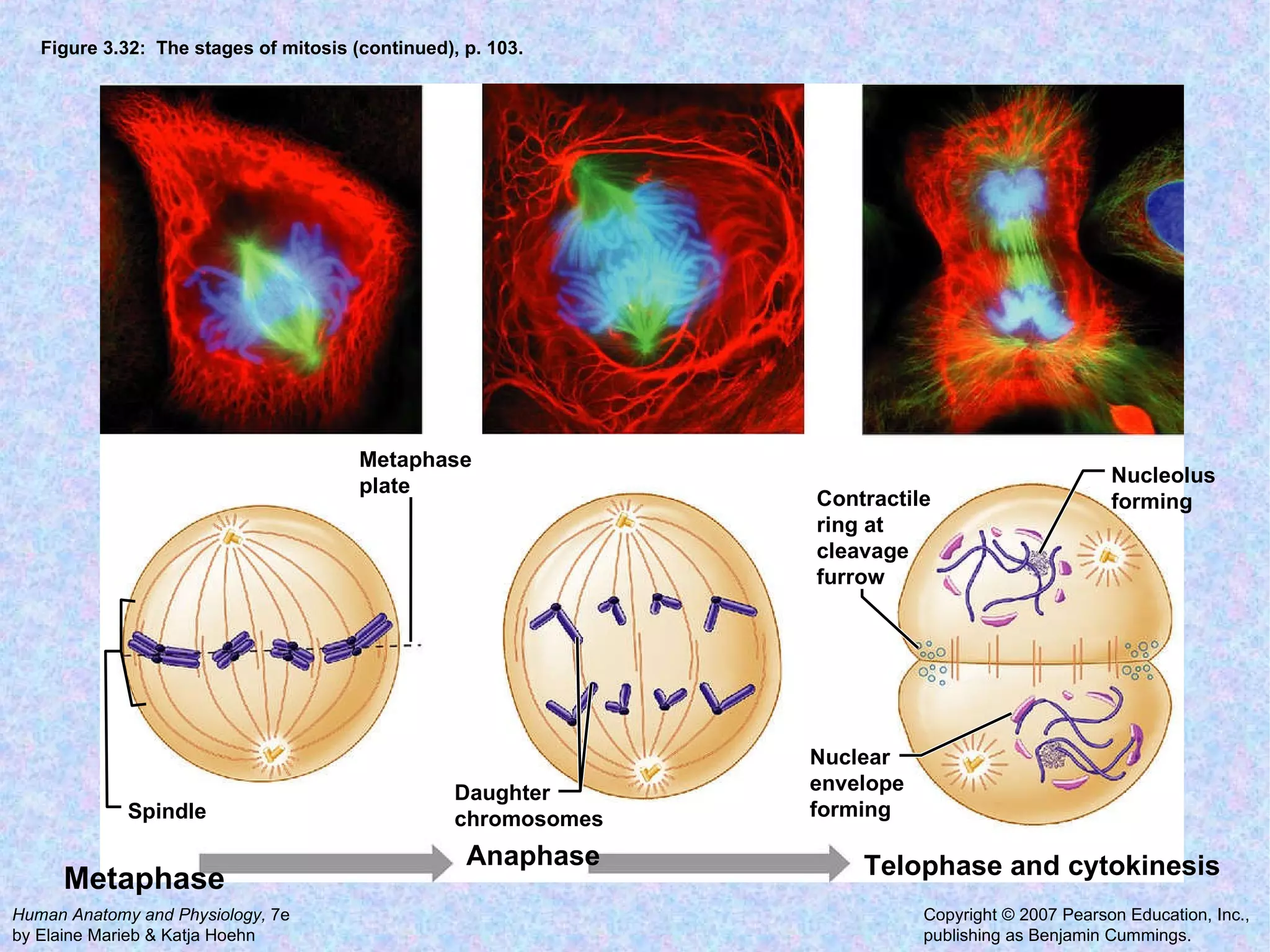
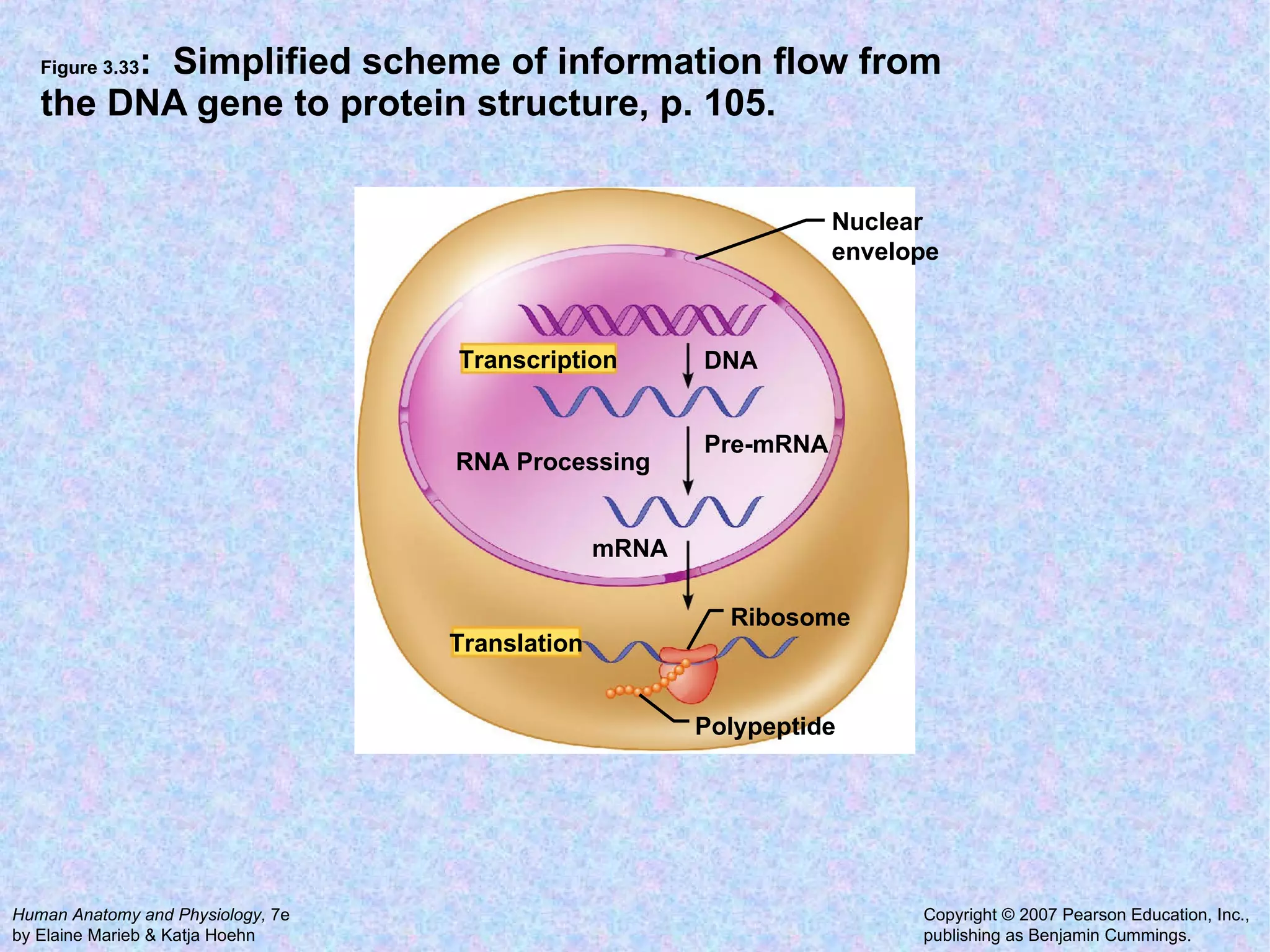
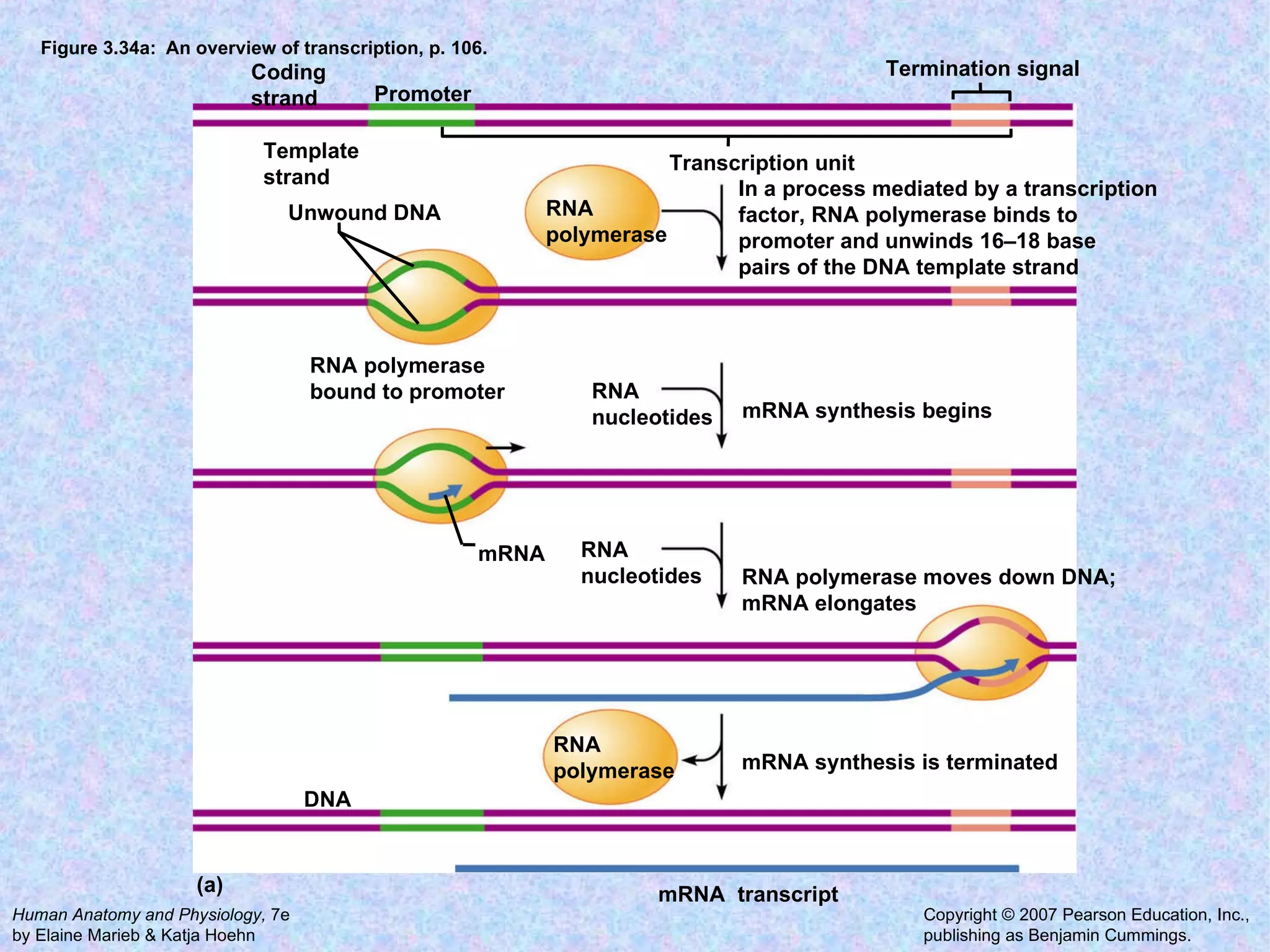
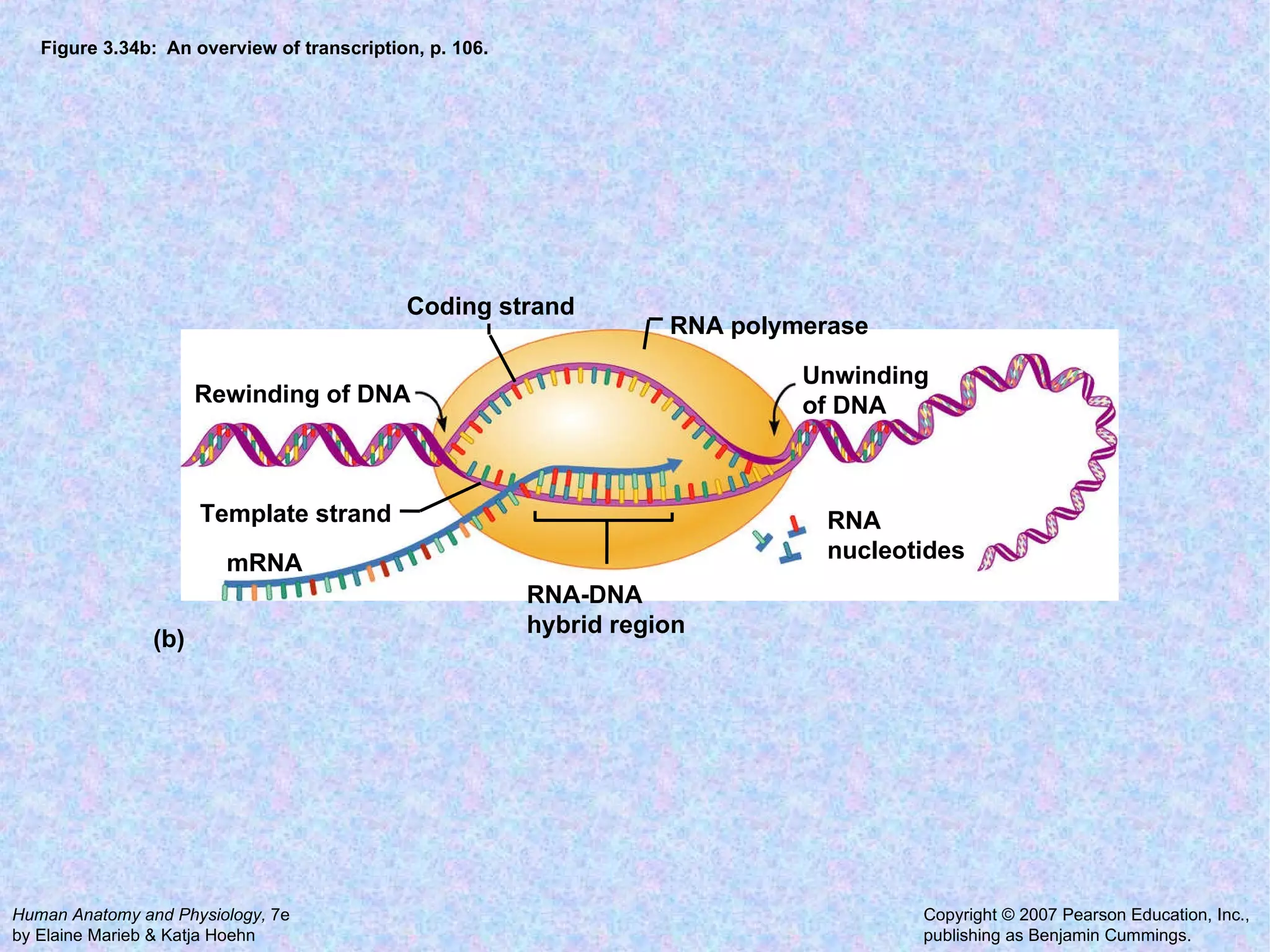
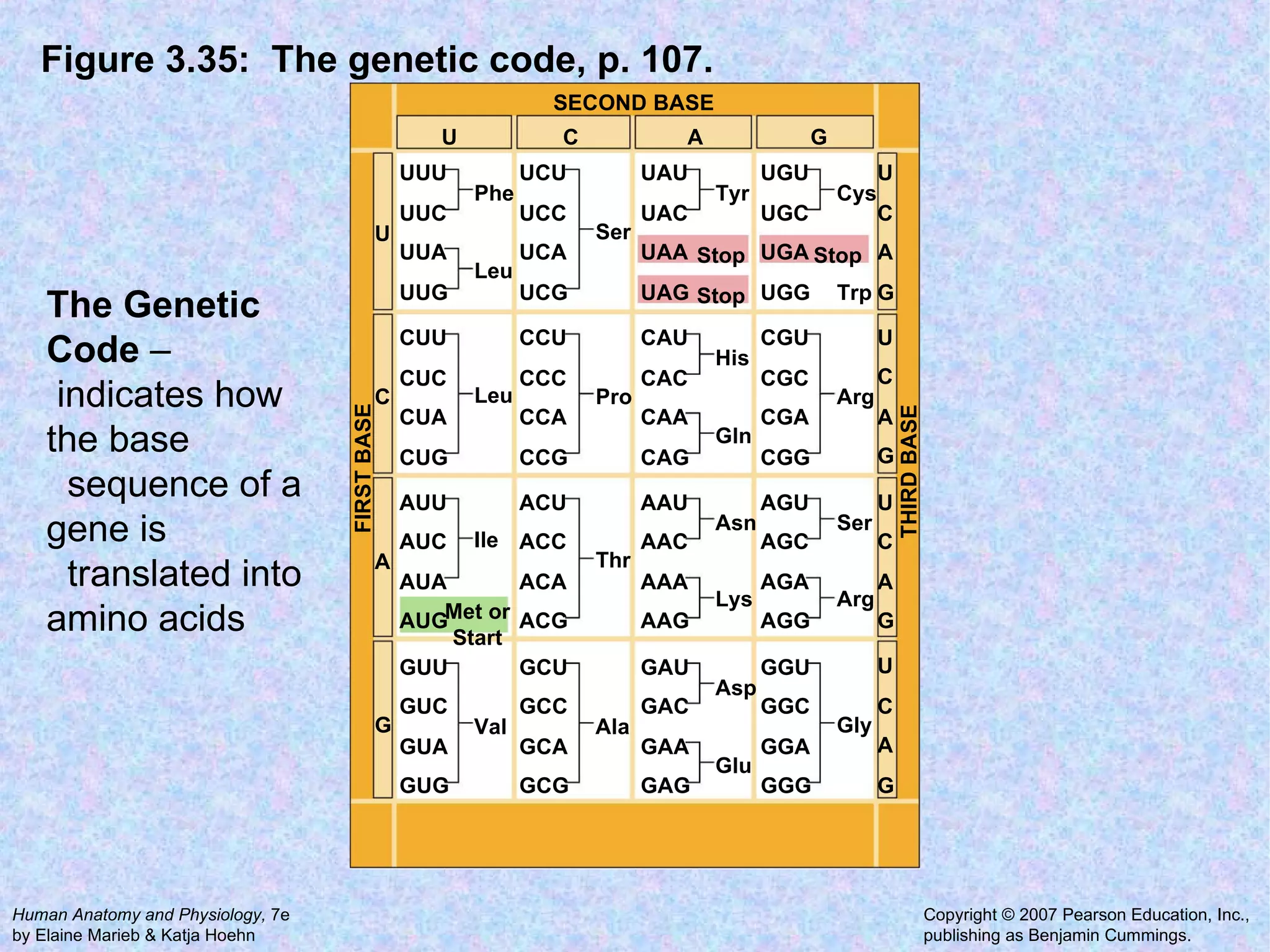
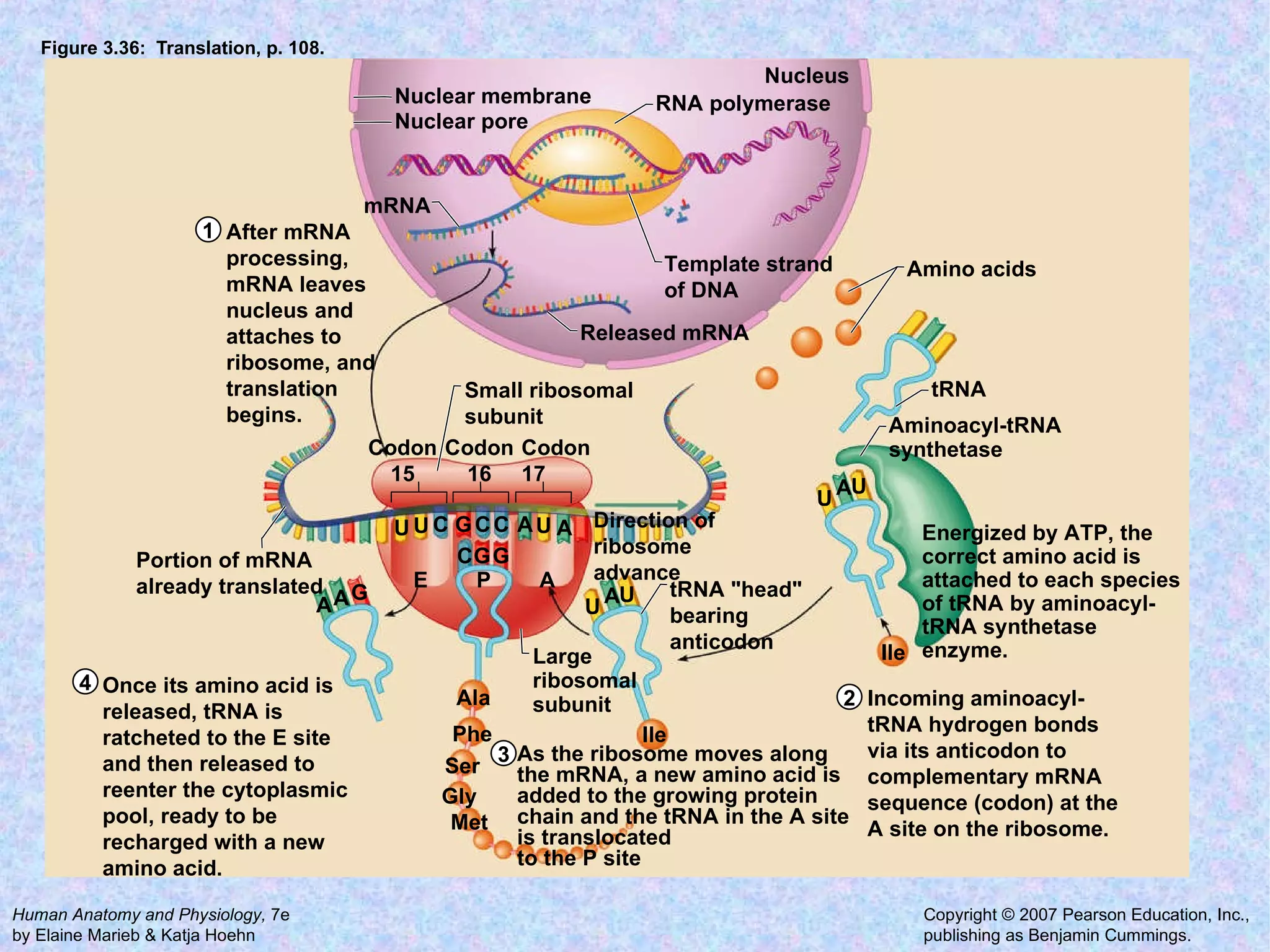
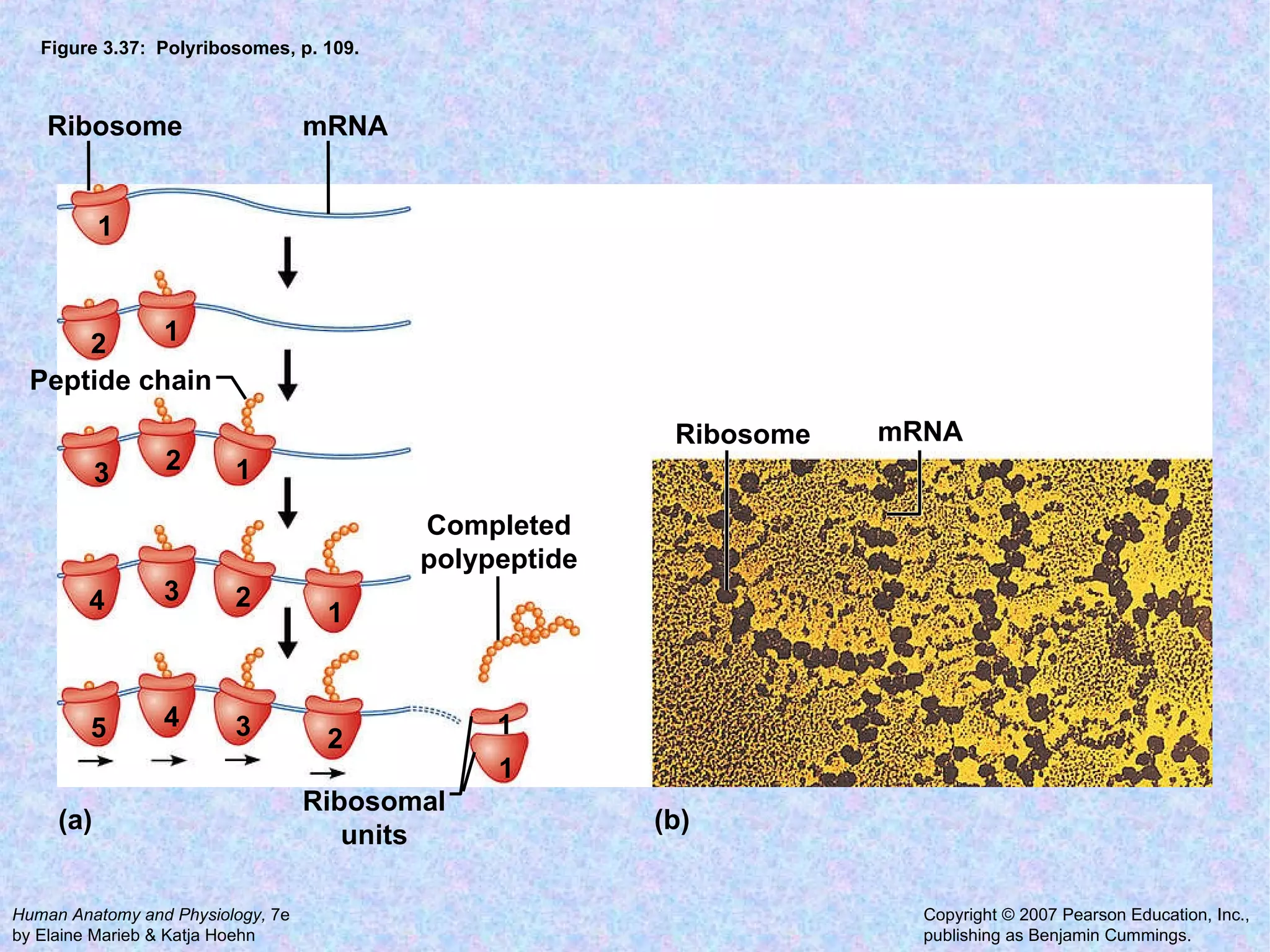
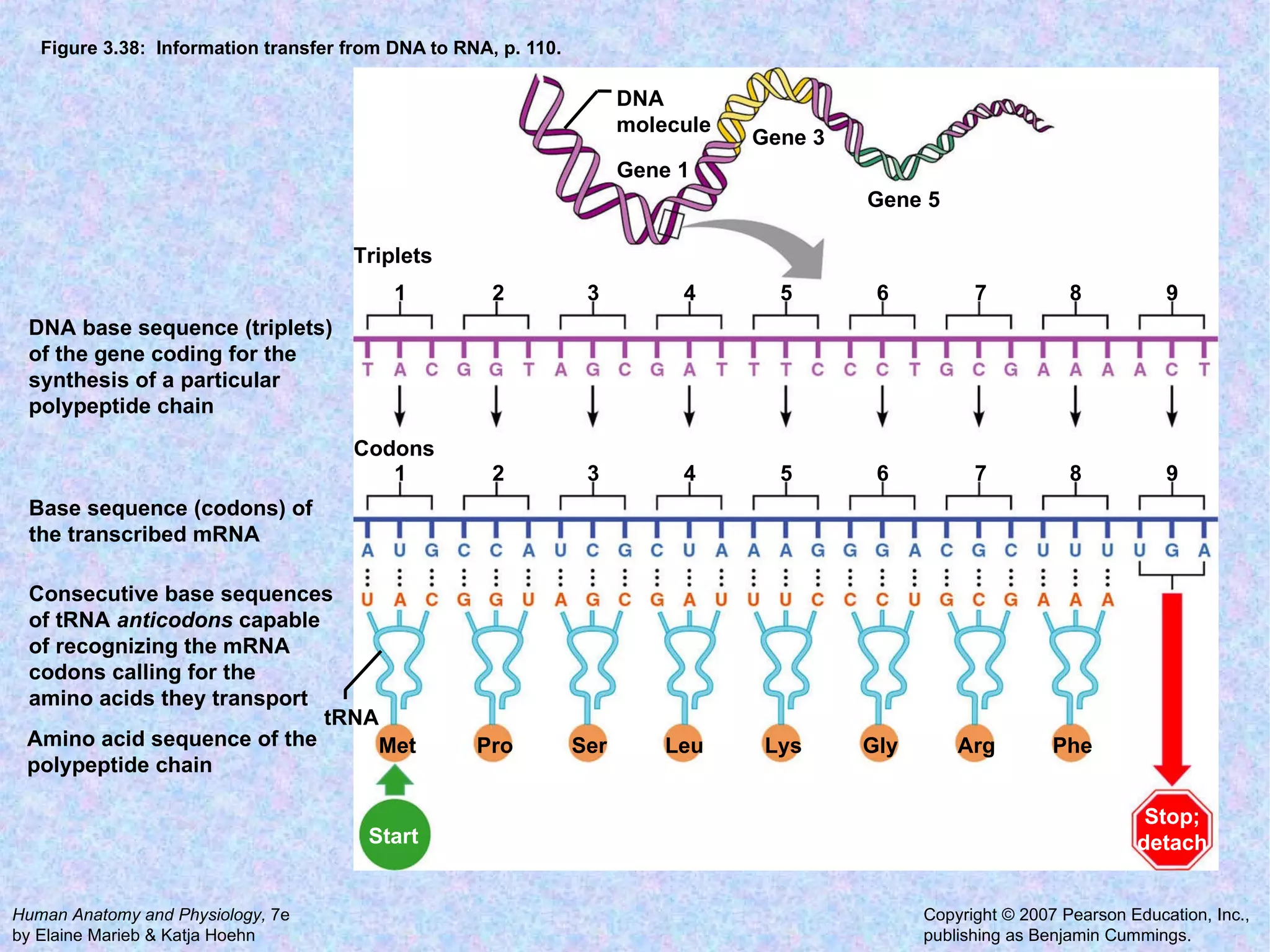
The document summarizes key concepts about cells:
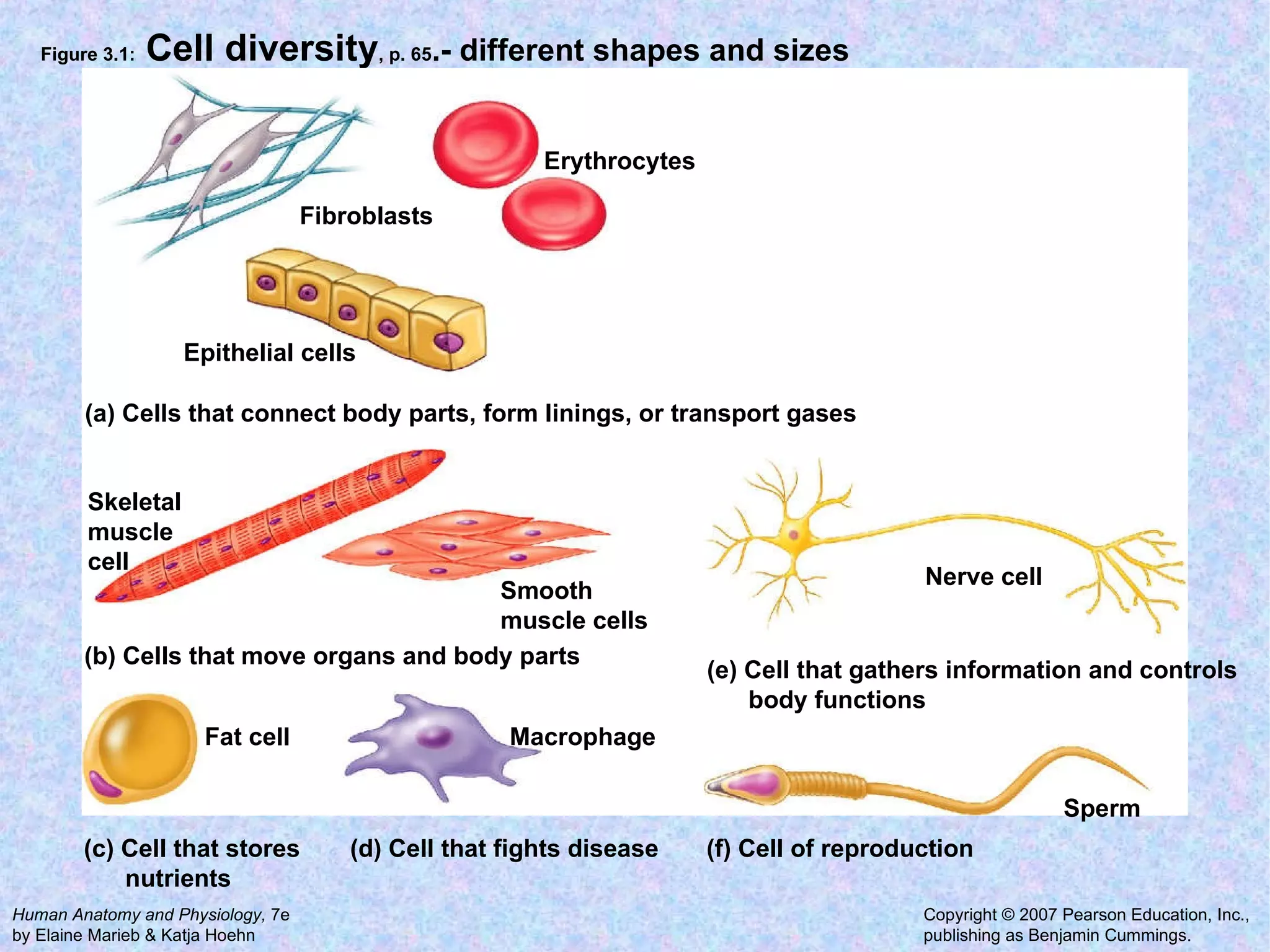
1. A cell is the basic structural and functional unit of all organisms. Cells come in different shapes and sizes and perform specialized functions.
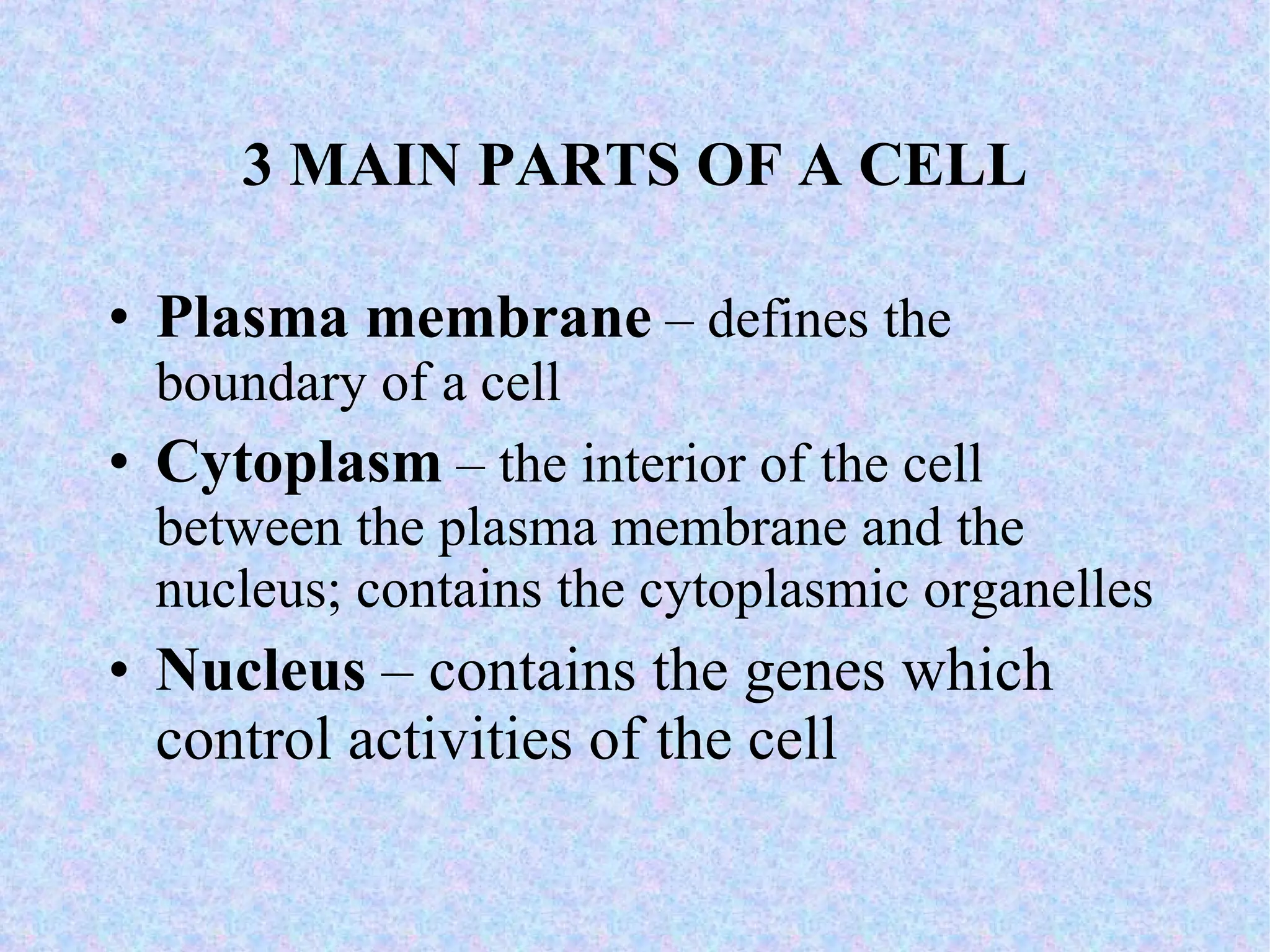
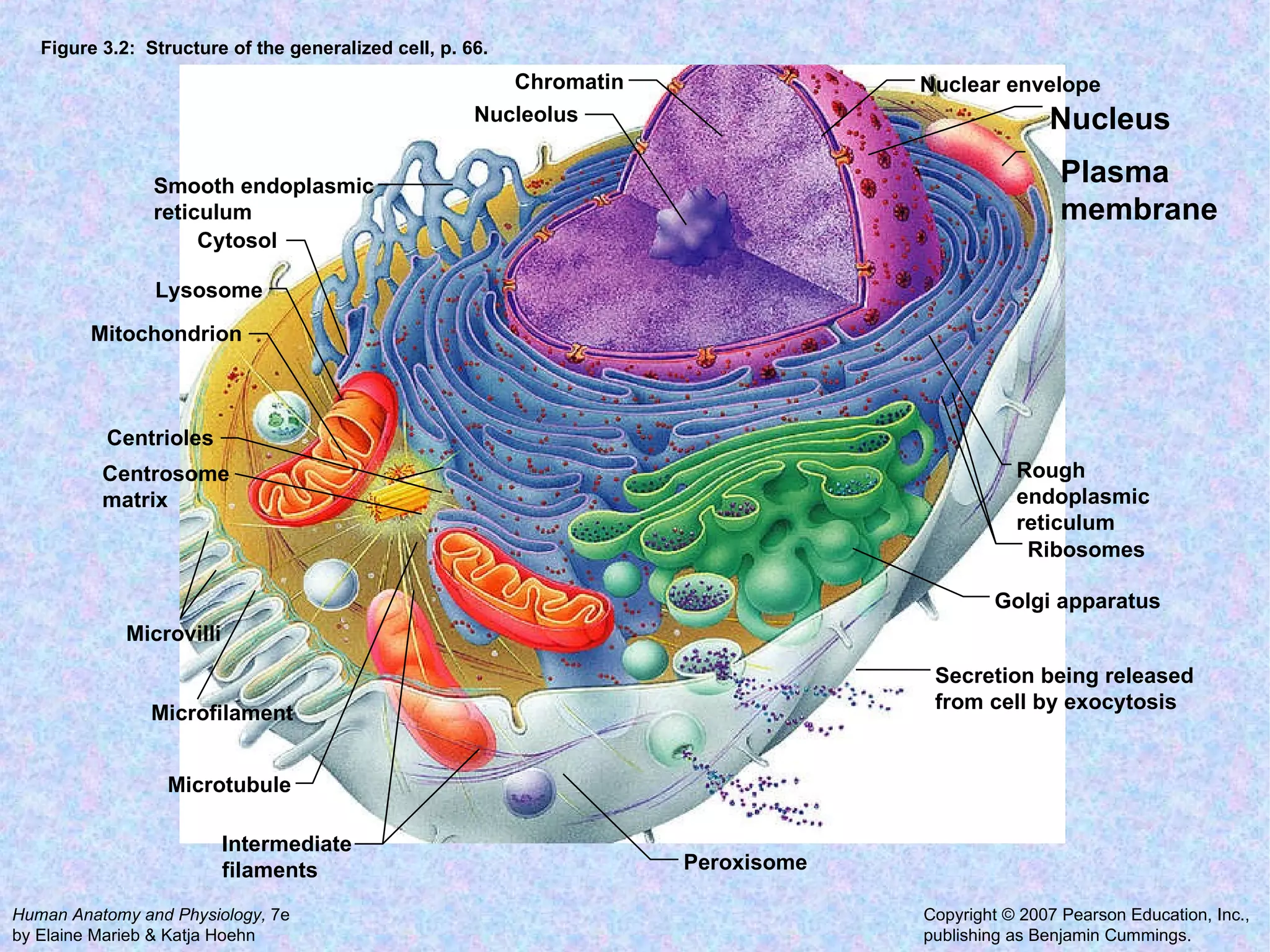
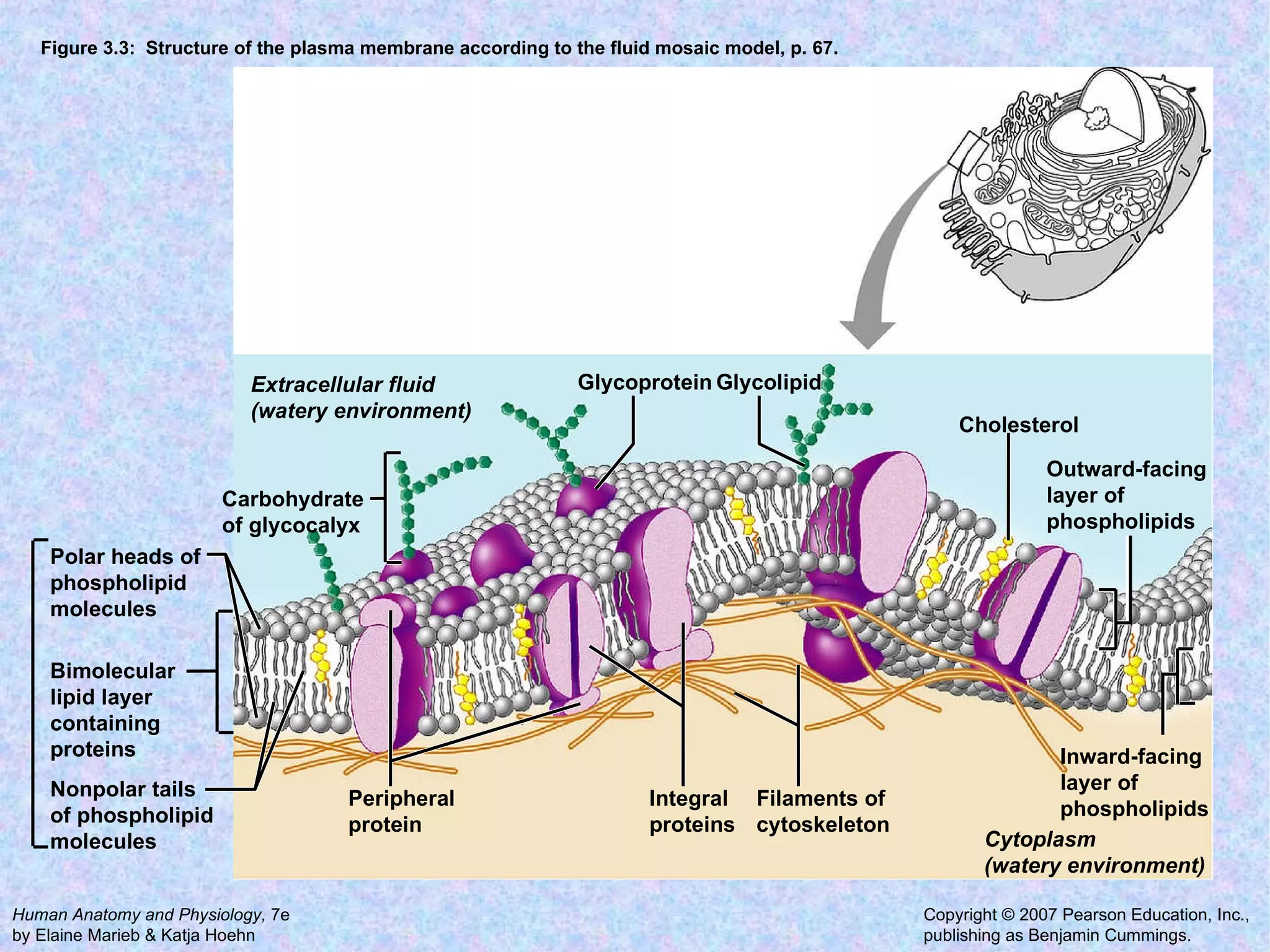
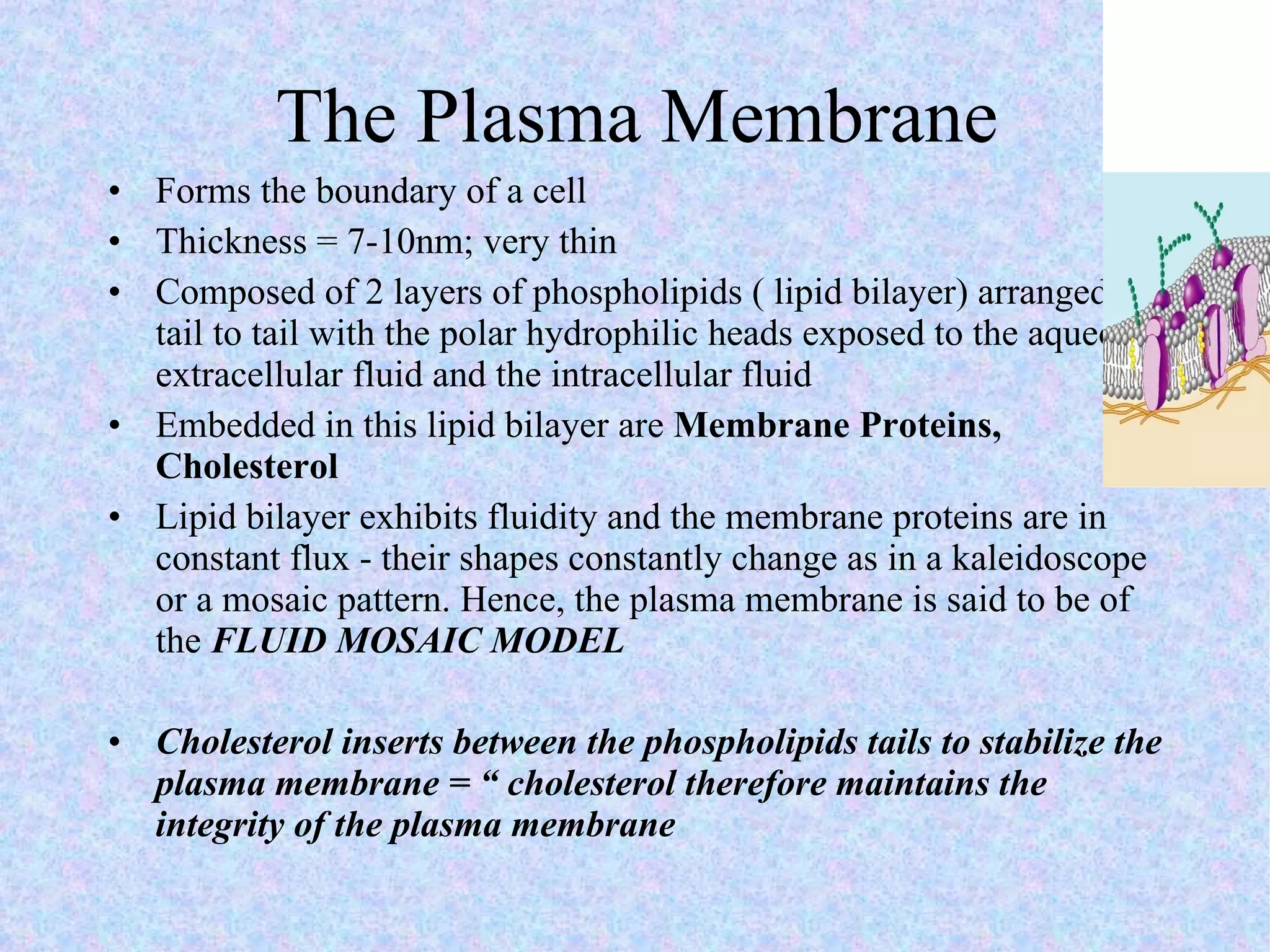
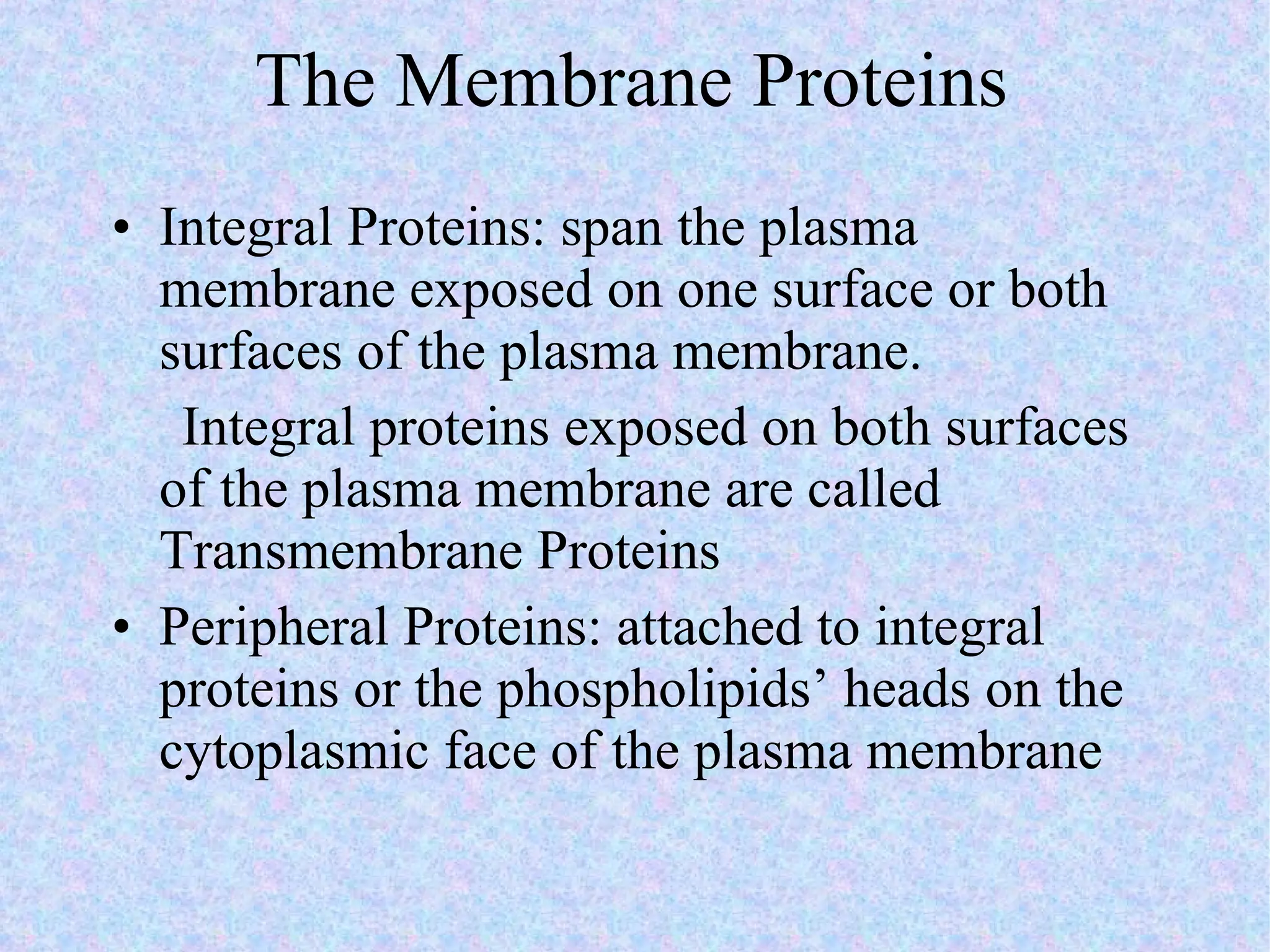
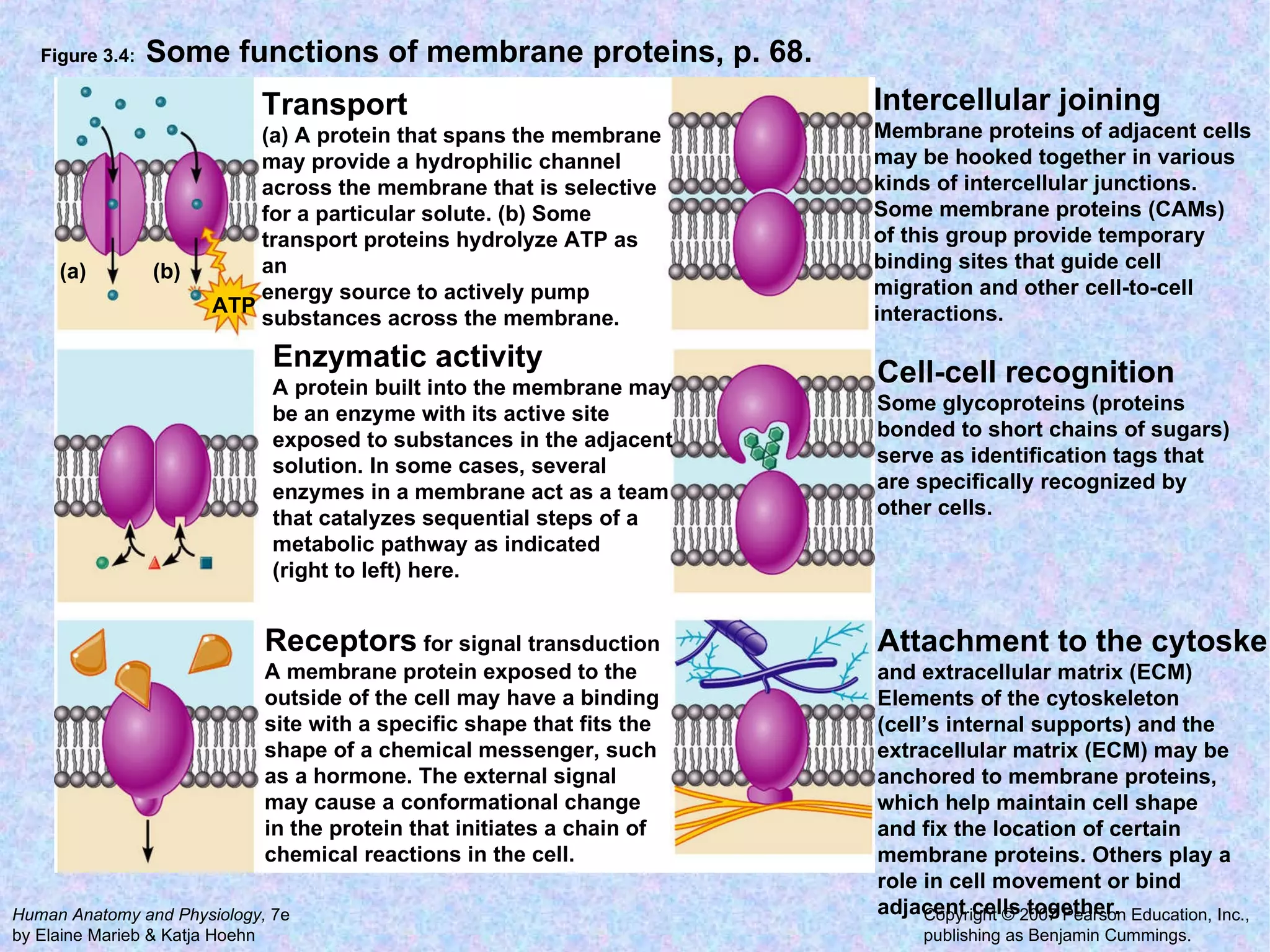
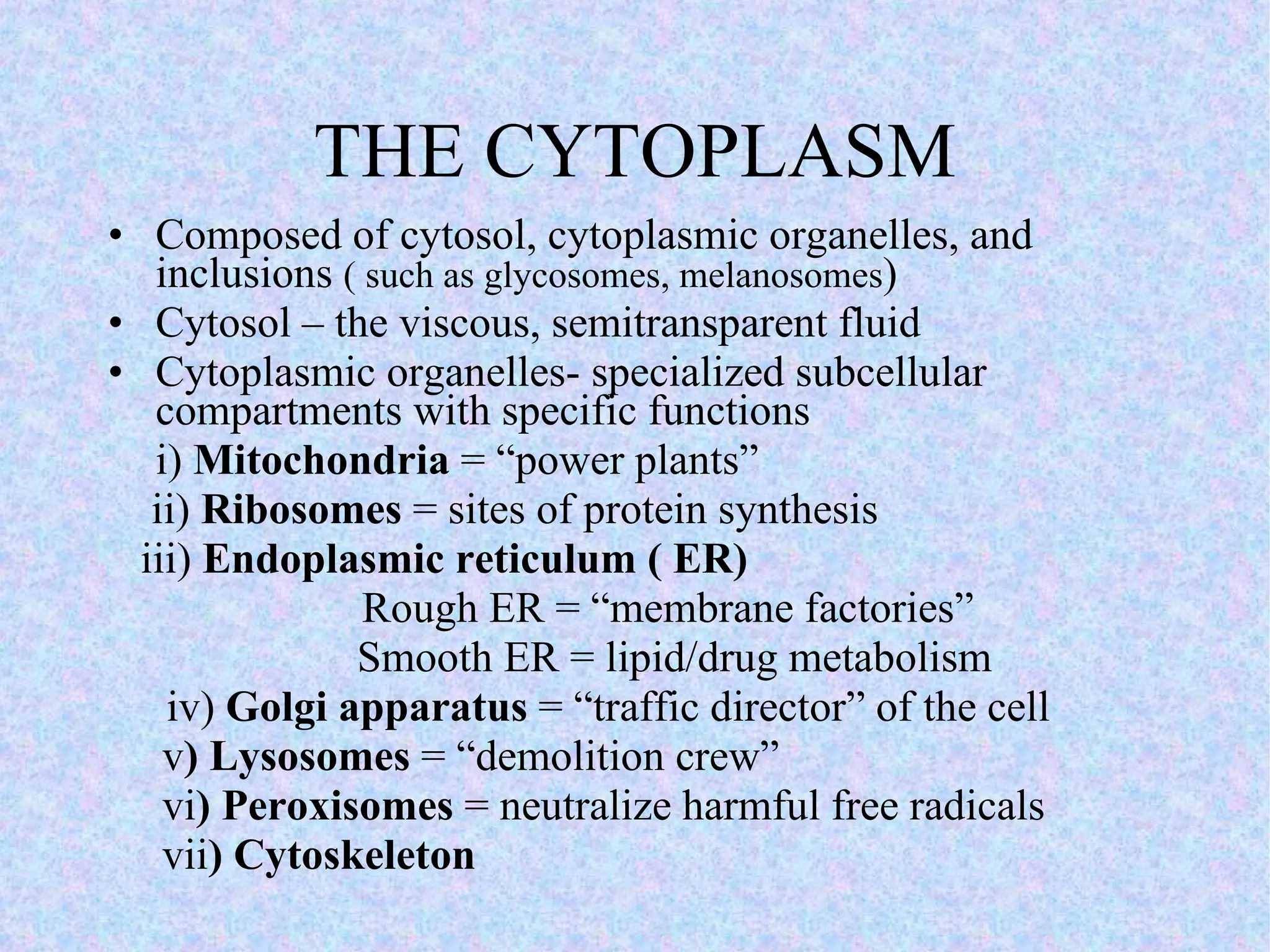
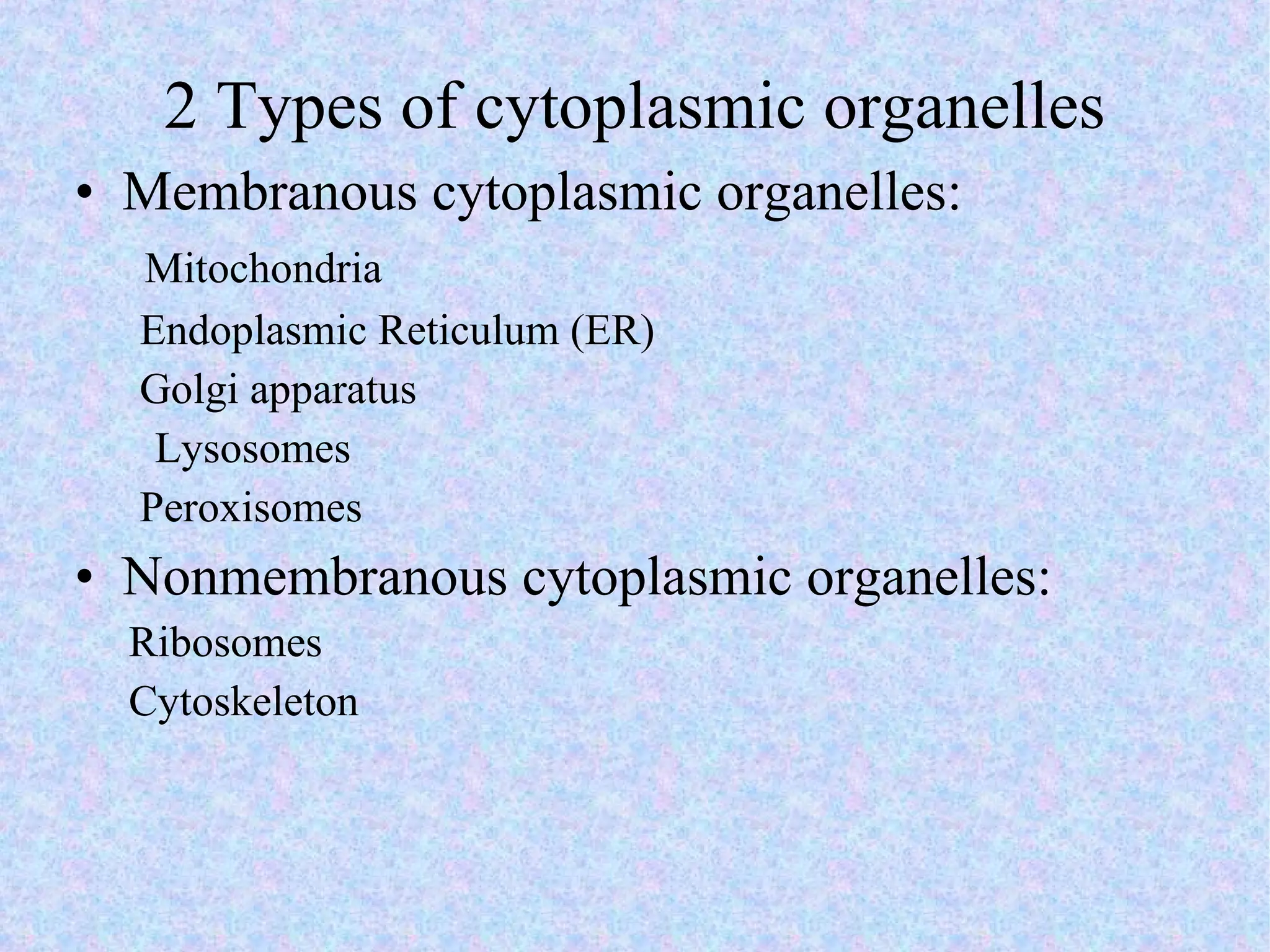
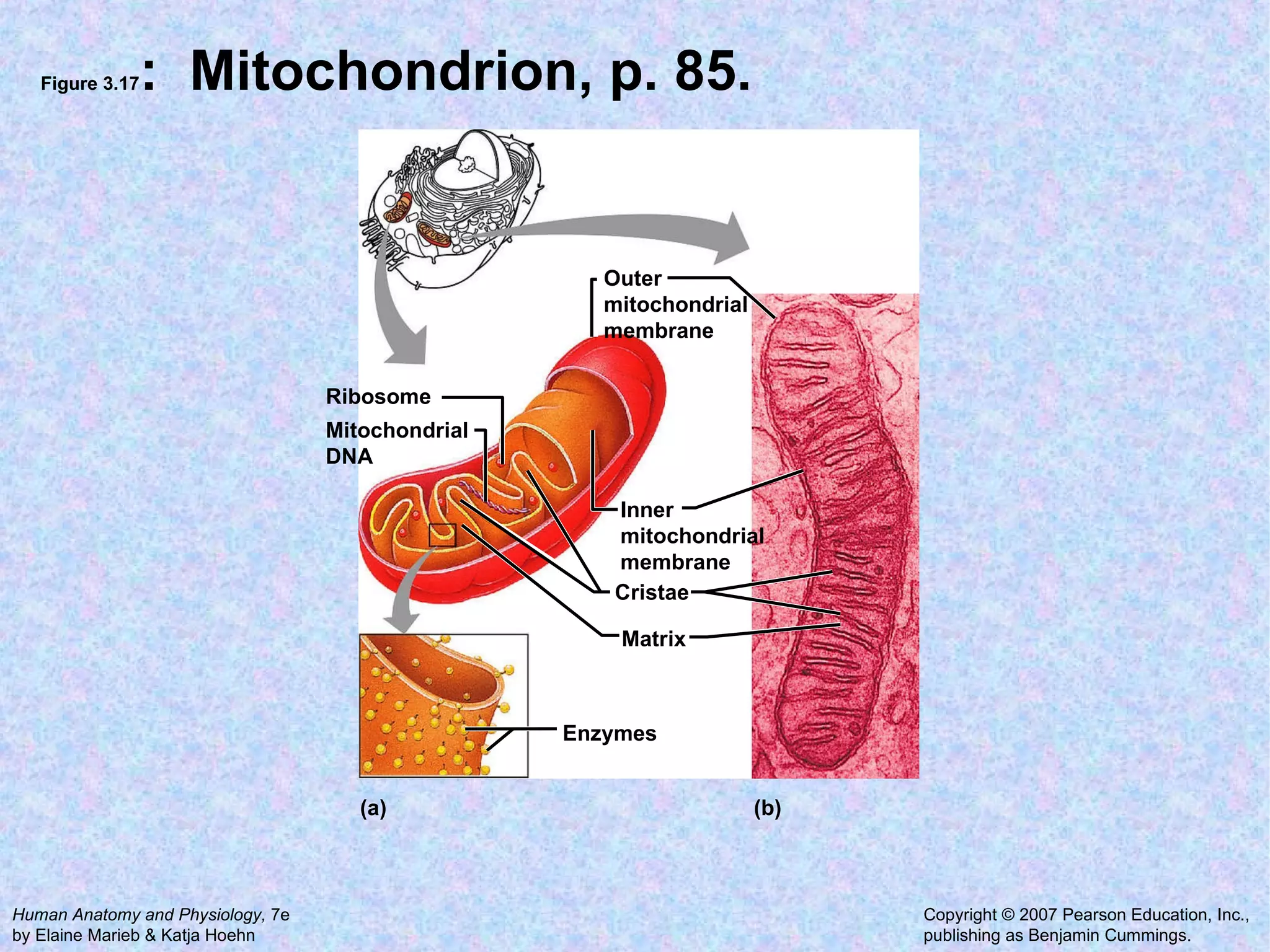
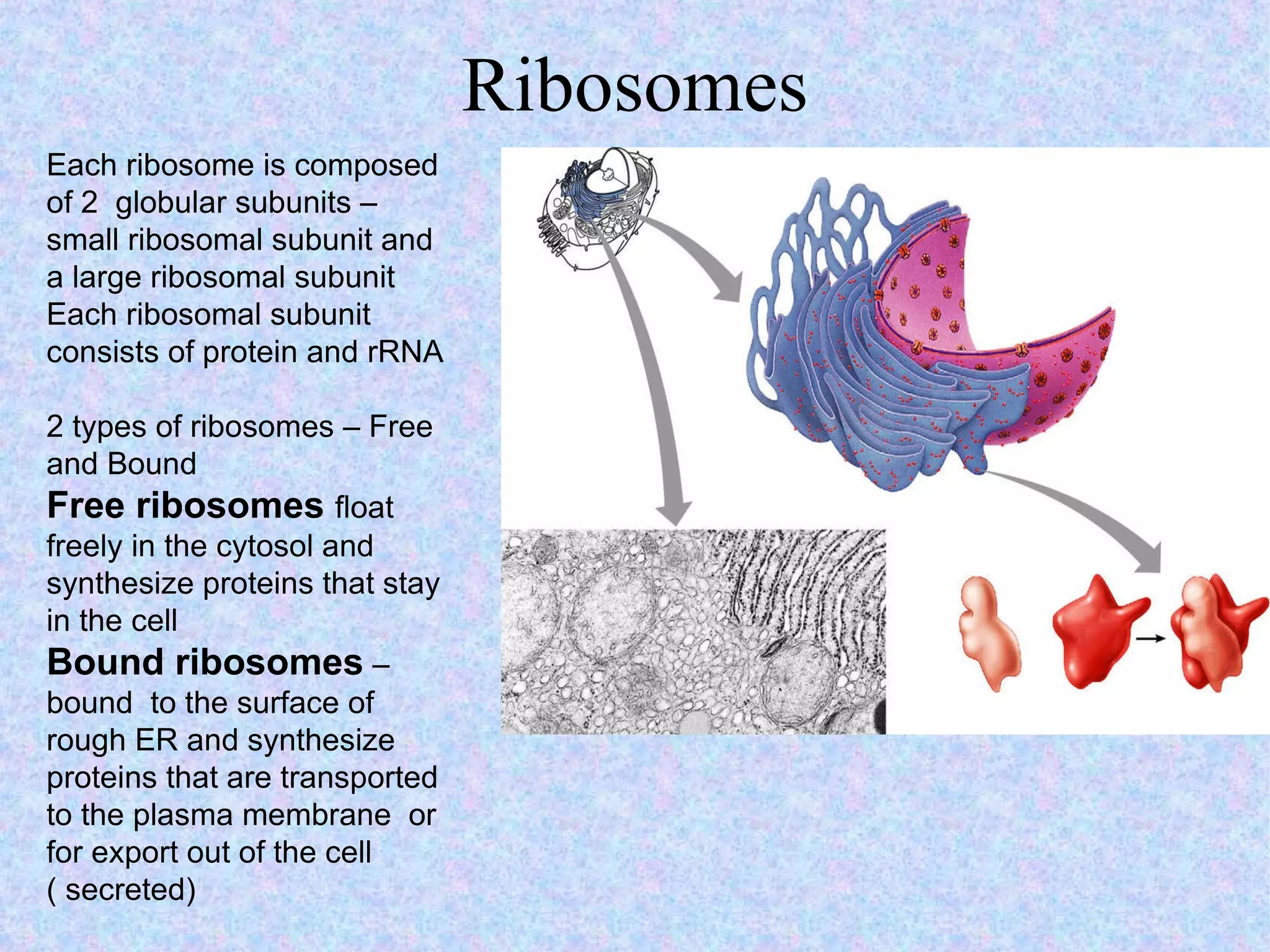
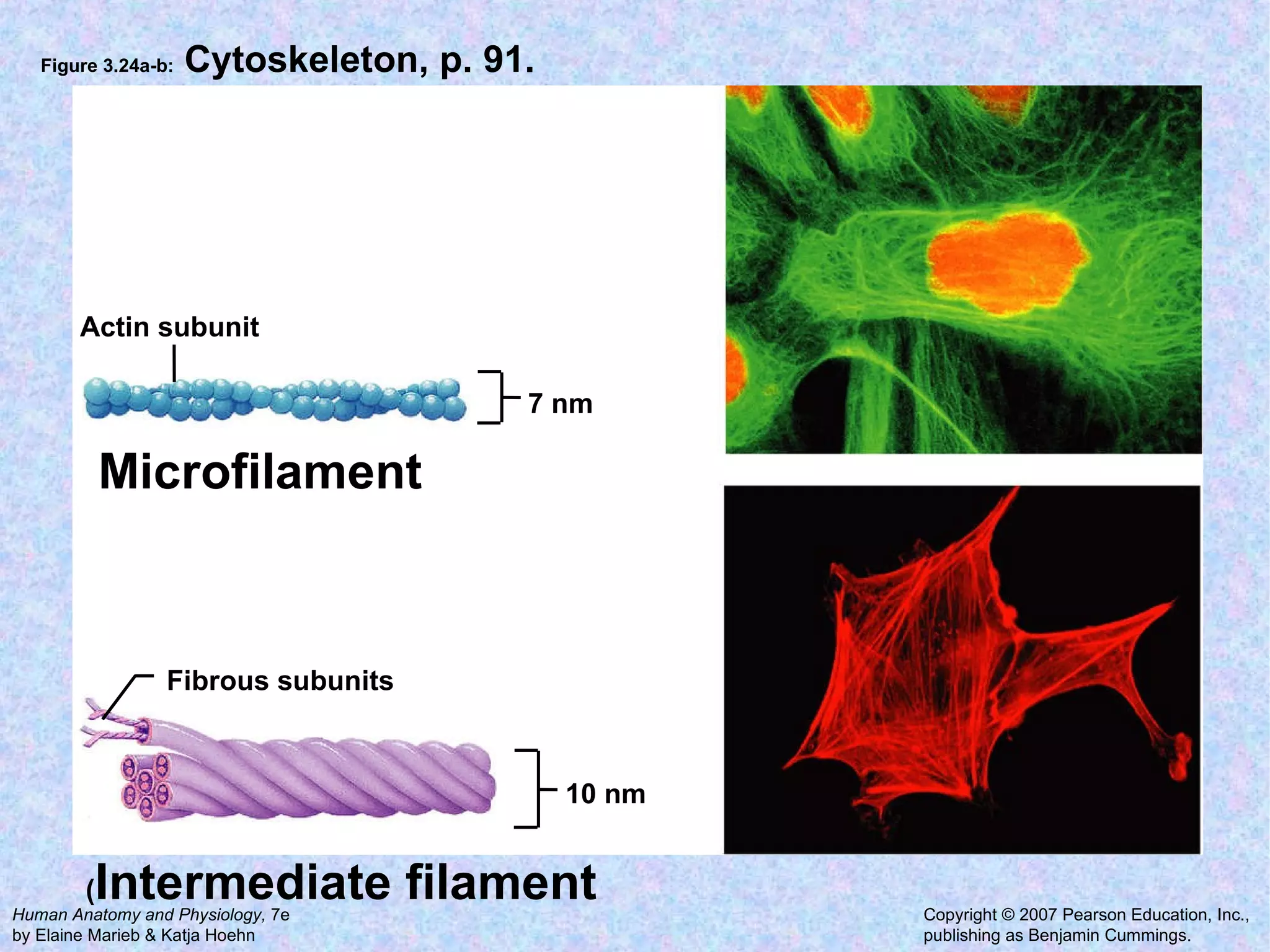
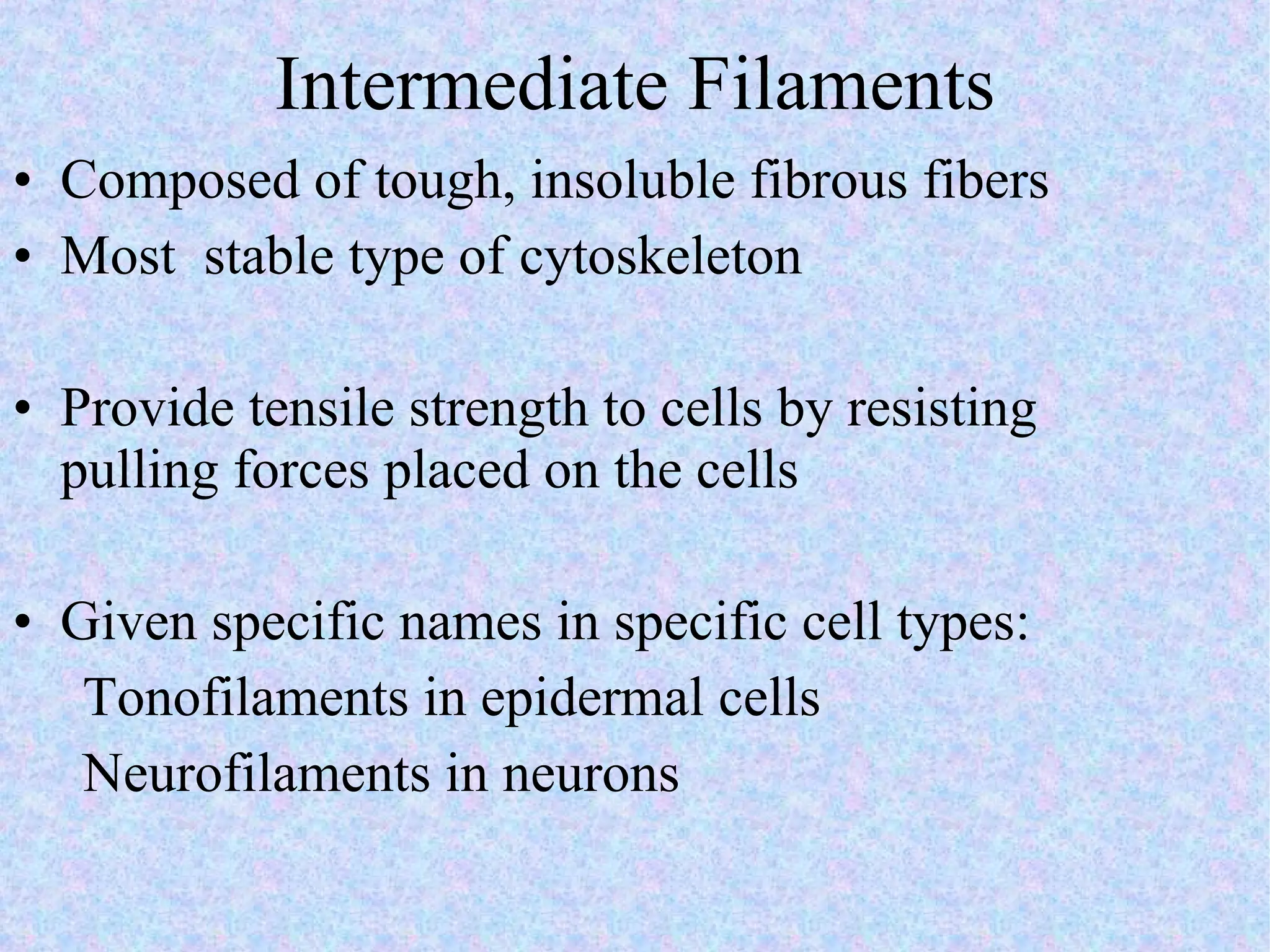
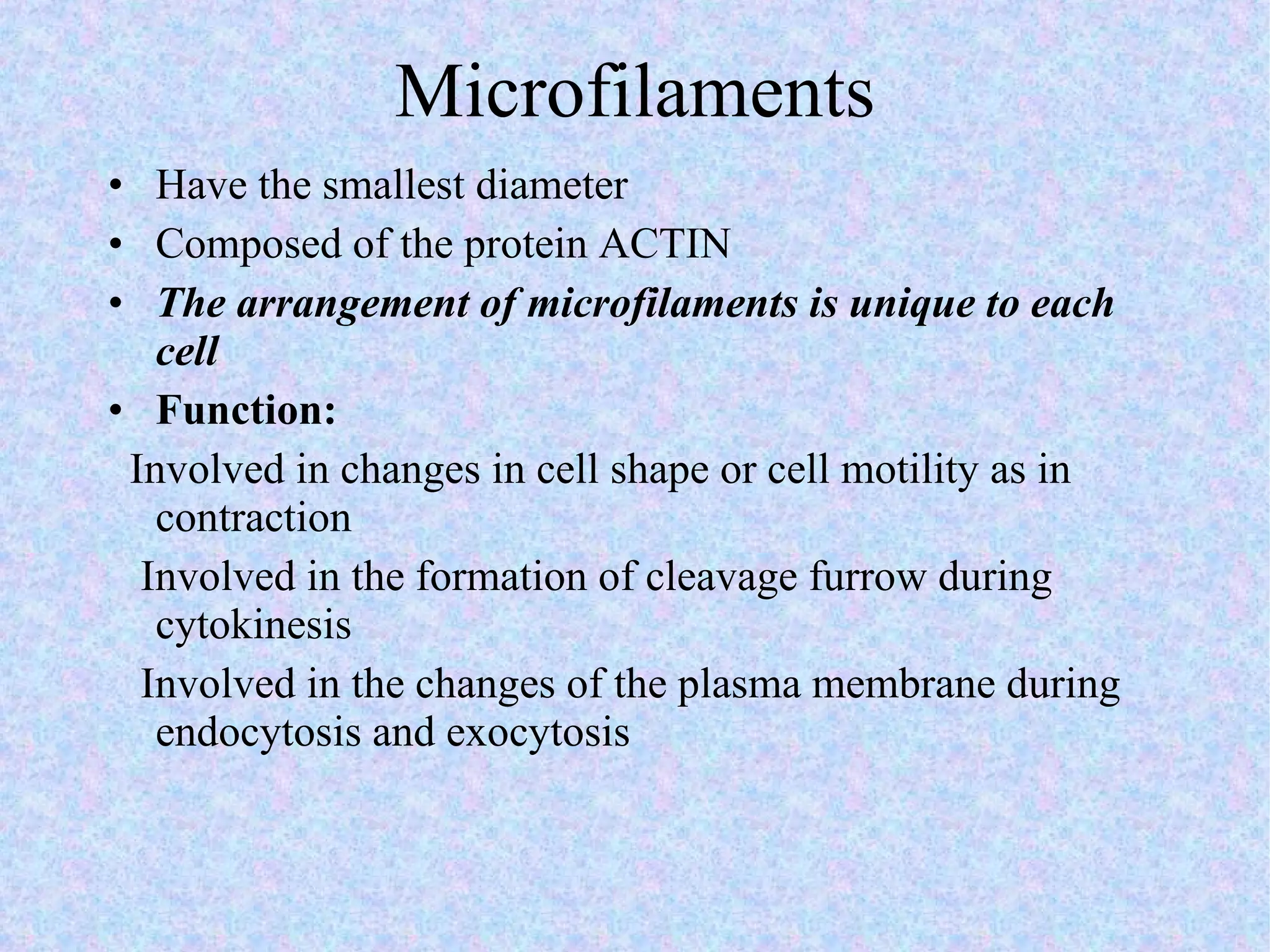
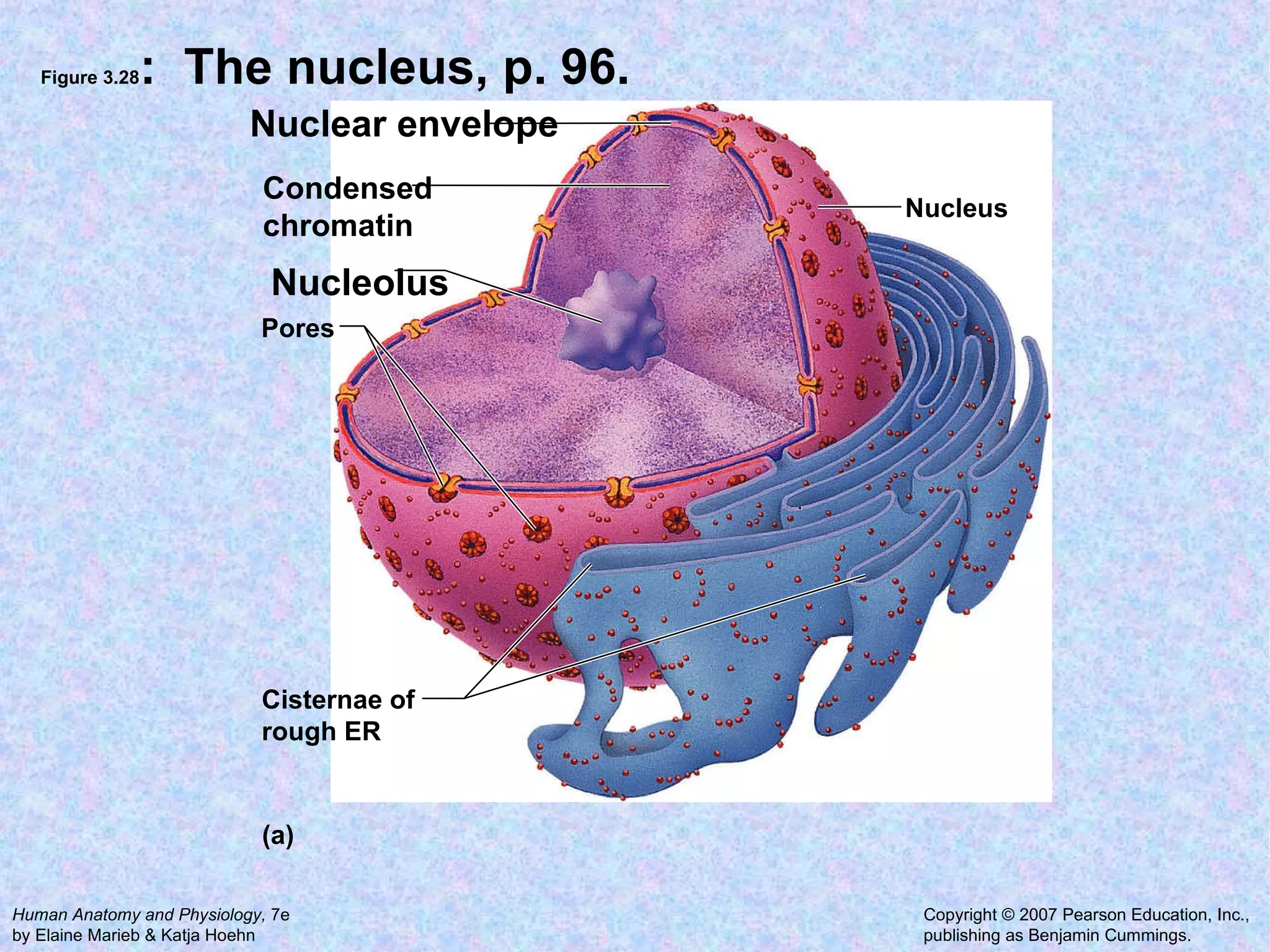
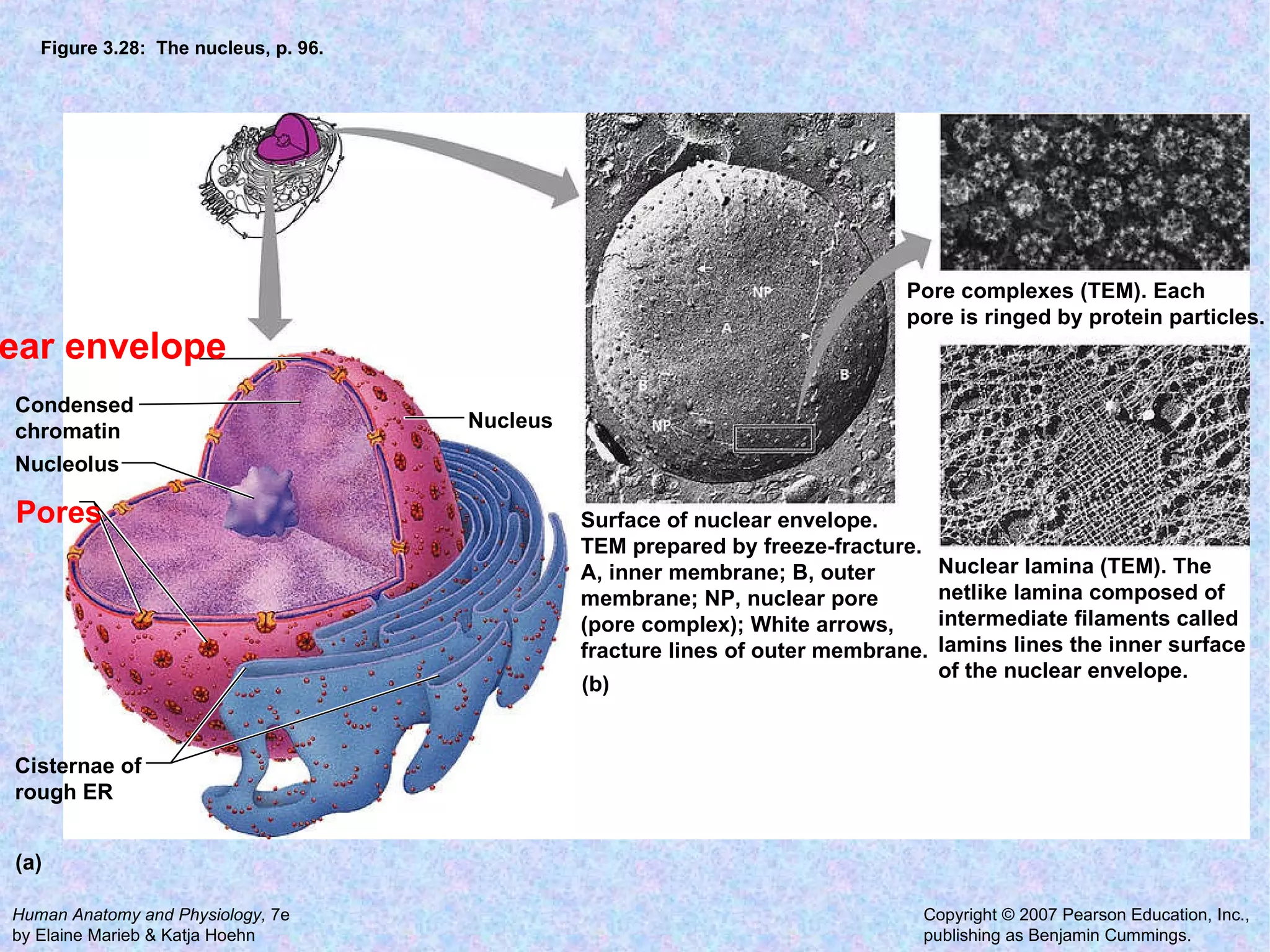
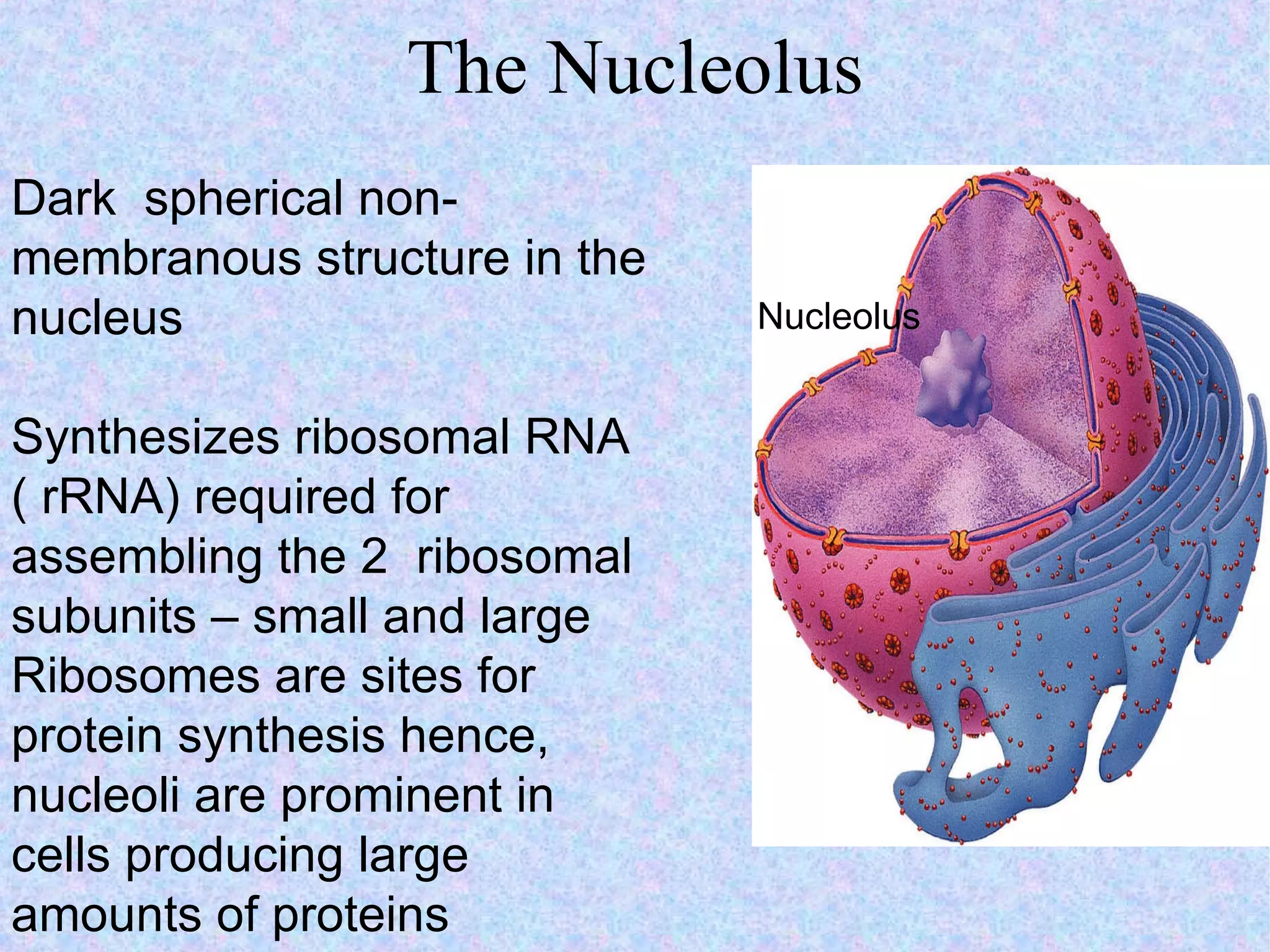
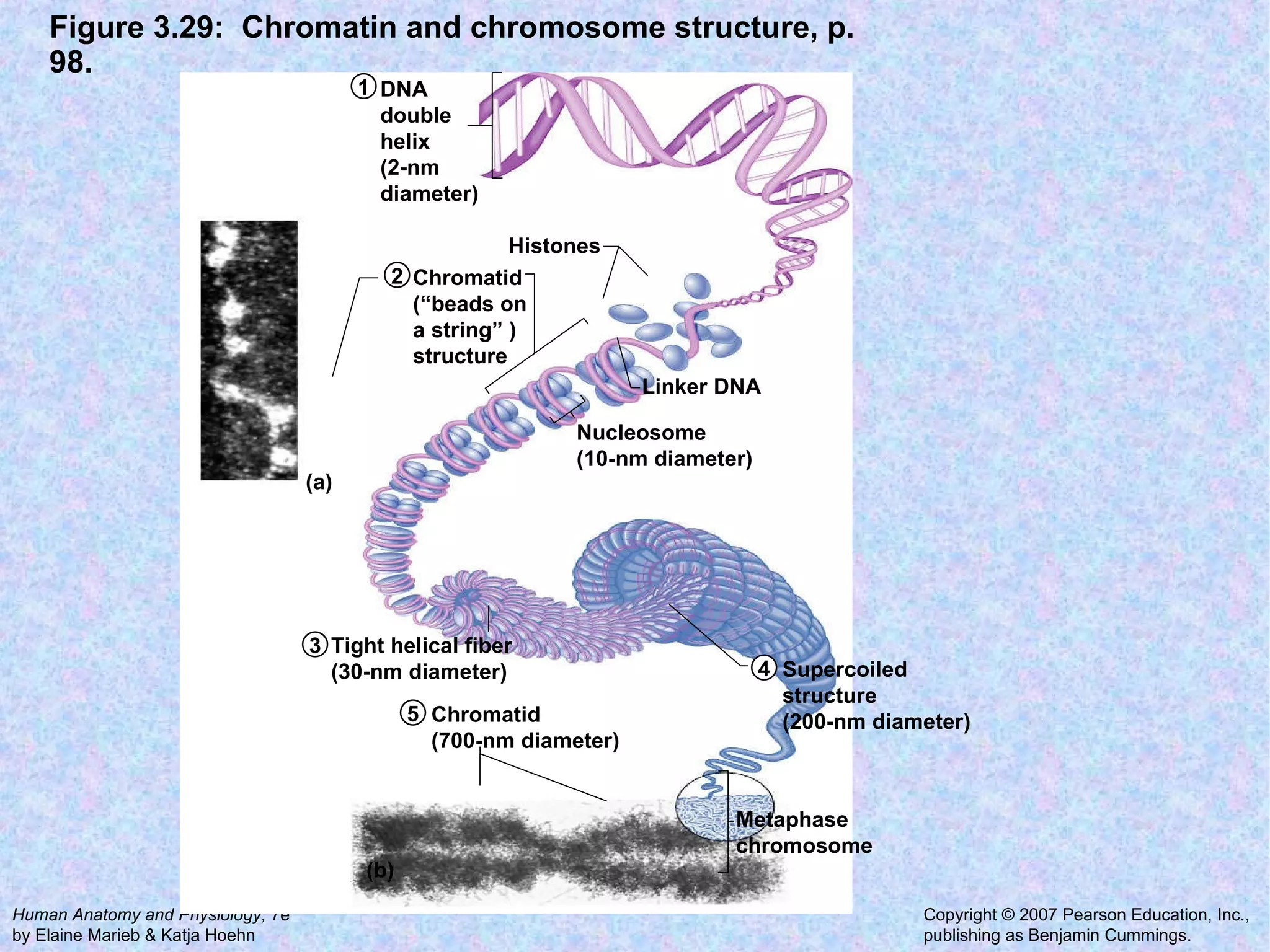
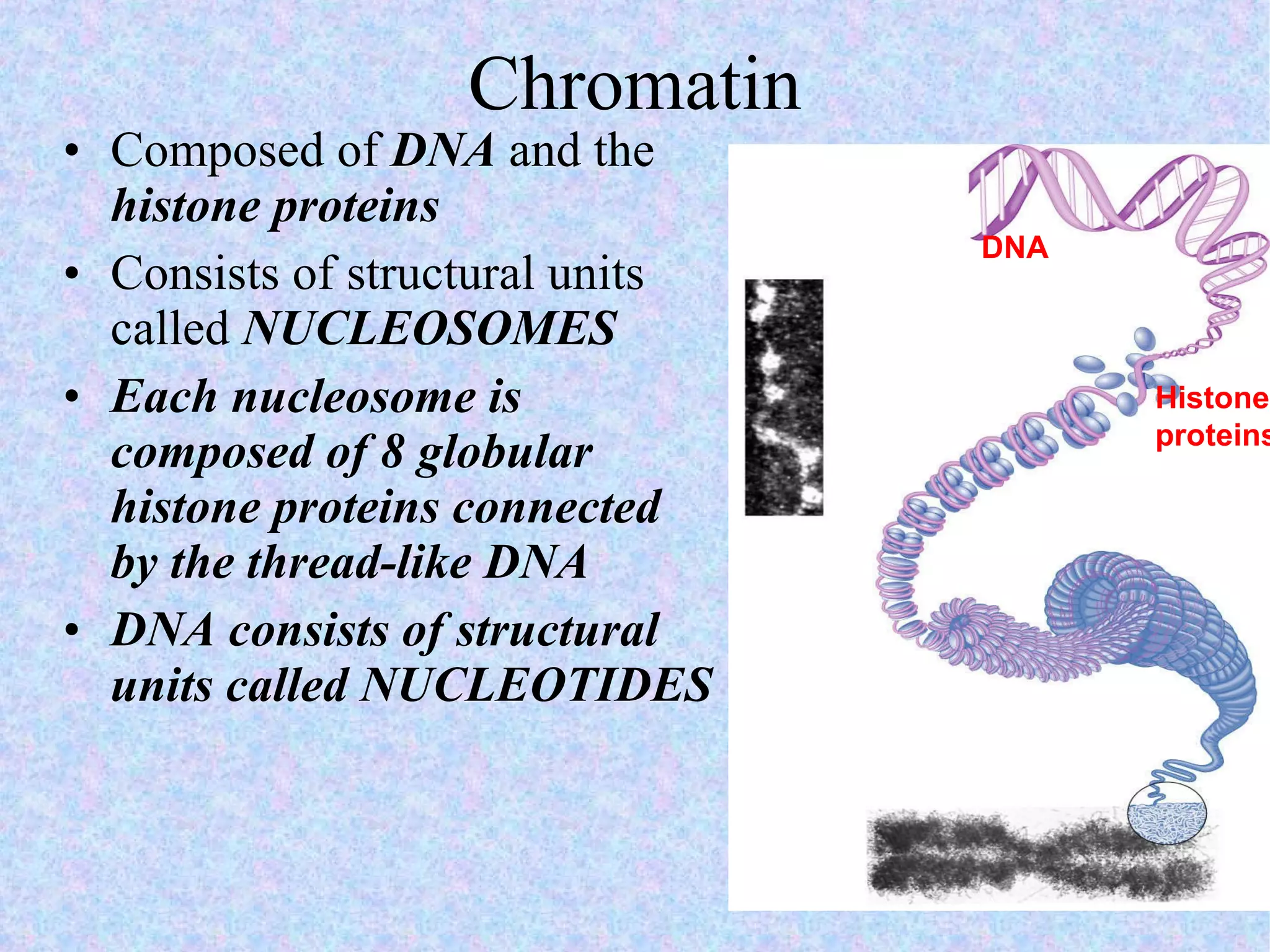
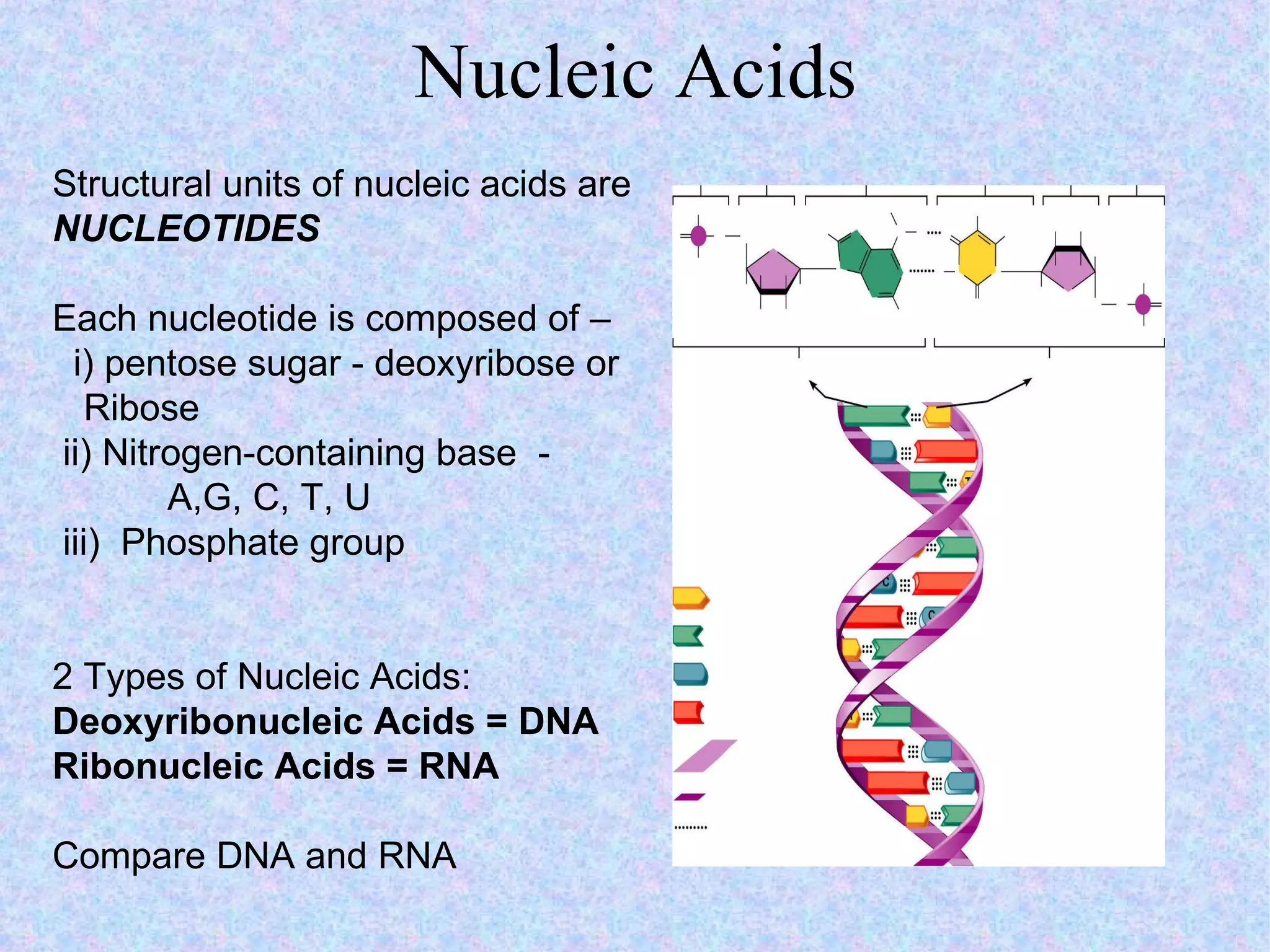
2. The main parts of a cell are the plasma membrane, cytoplasm, and nucleus. The plasma membrane defines the cell boundary, the cytoplasm is the fluid inside the cell, and the nucleus contains the cell's genes.
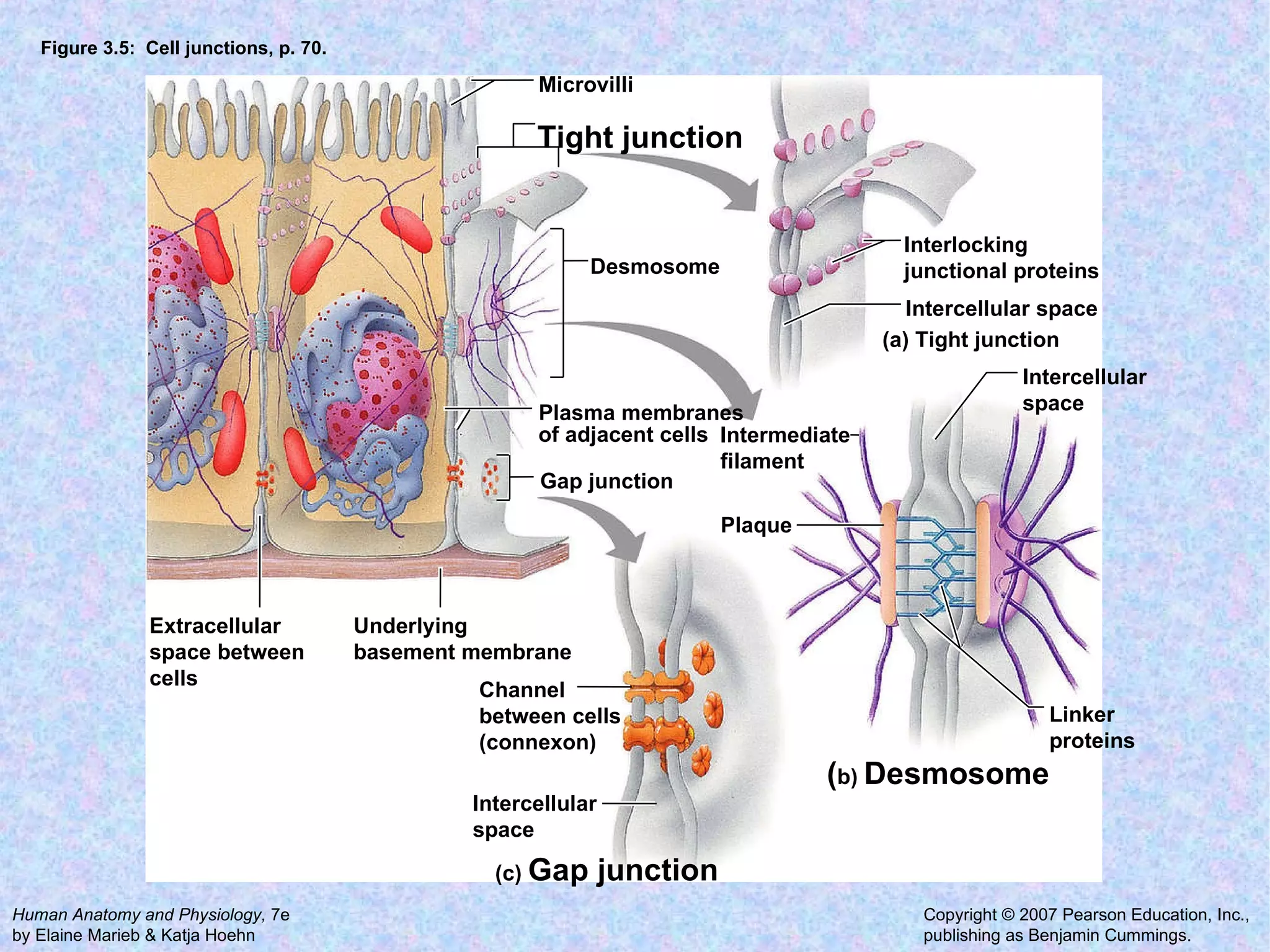
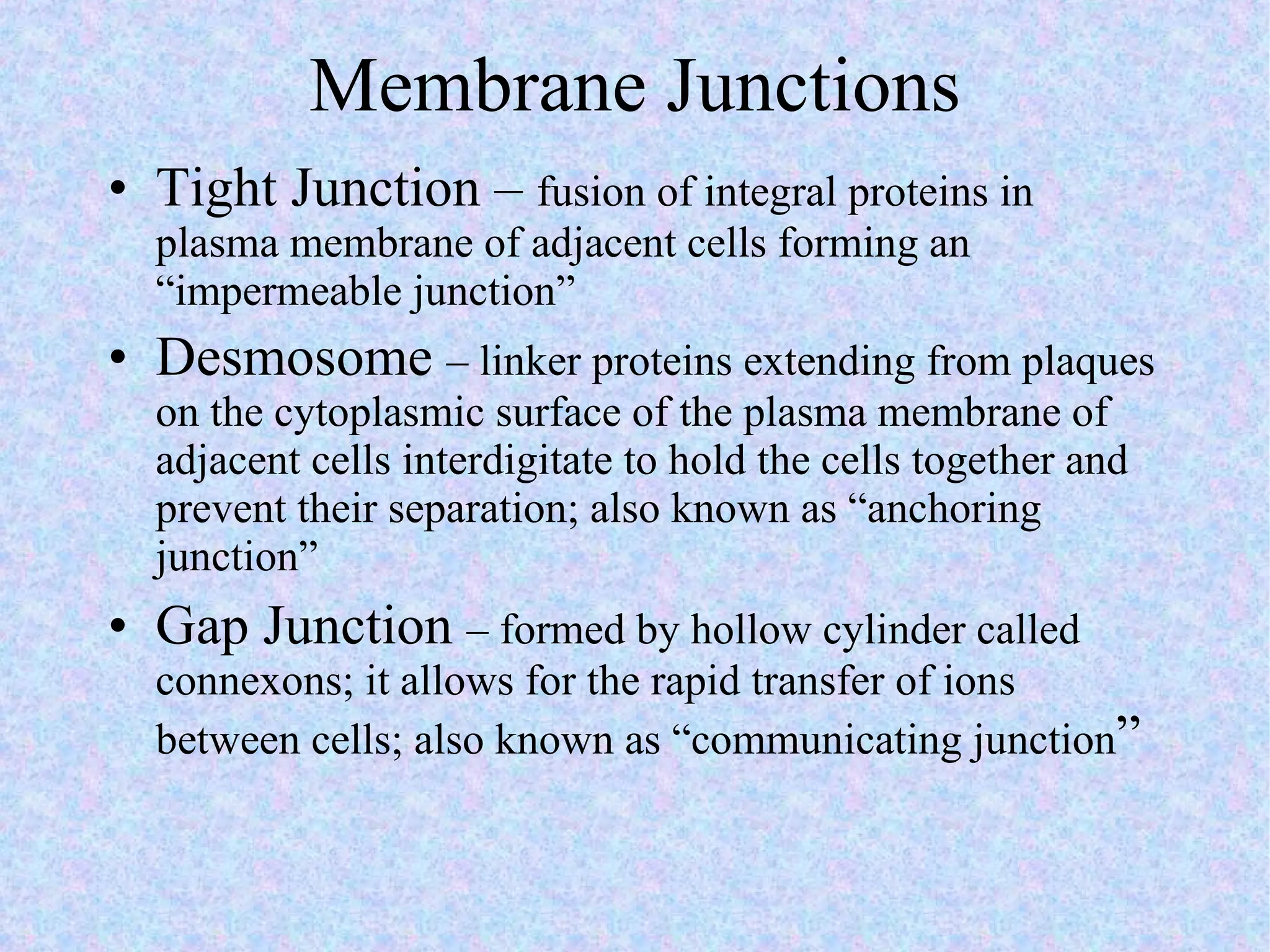
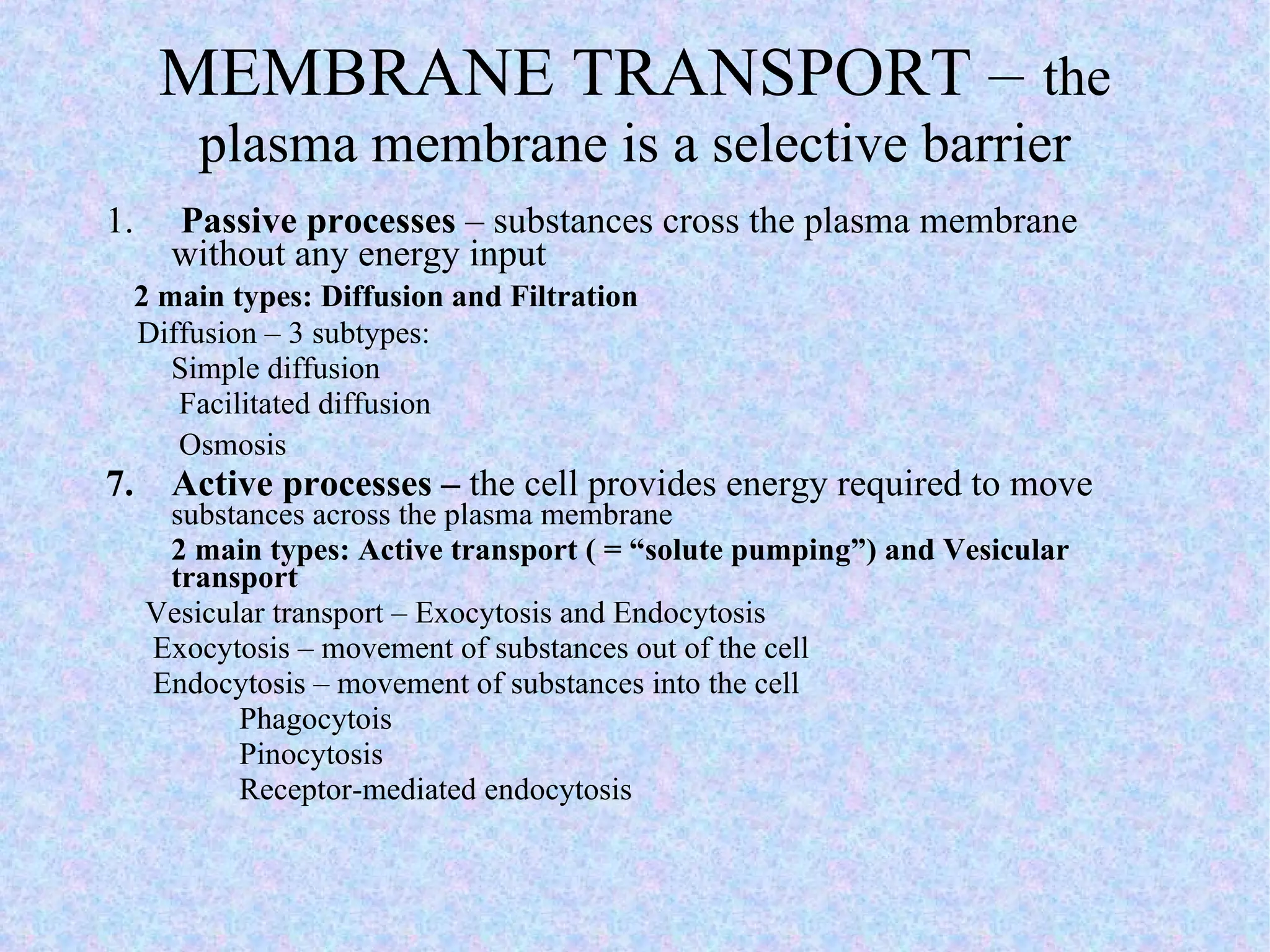
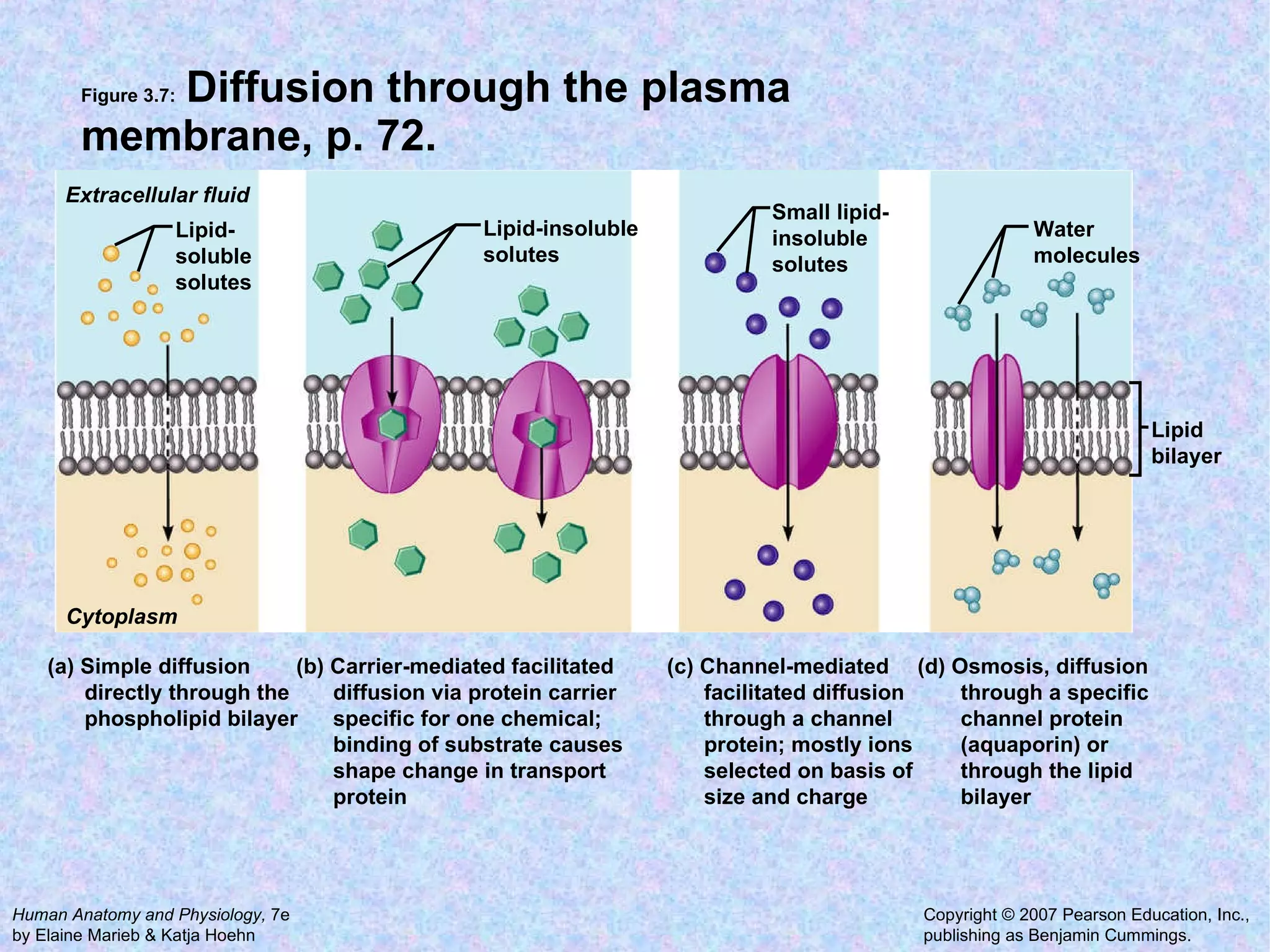
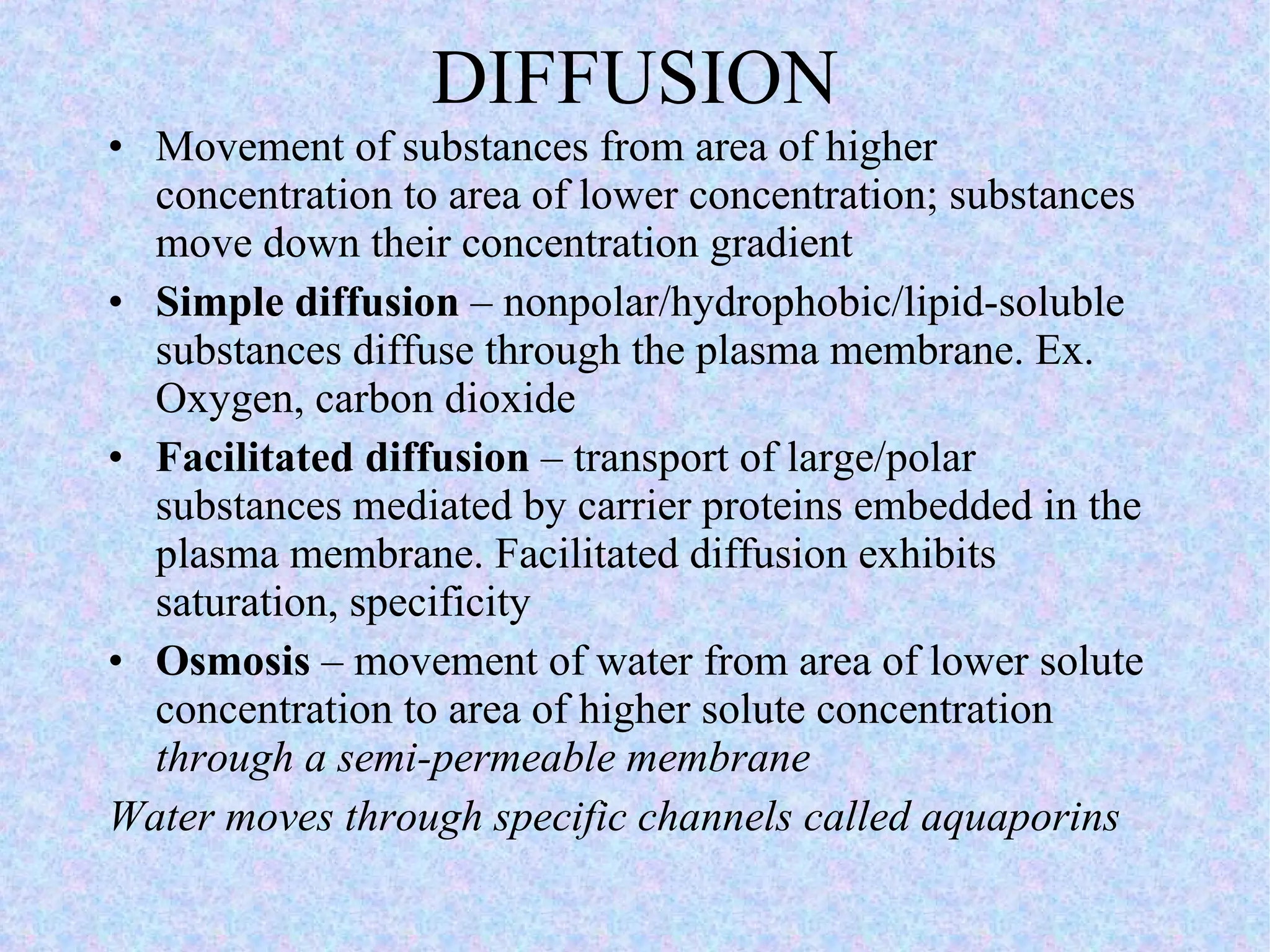
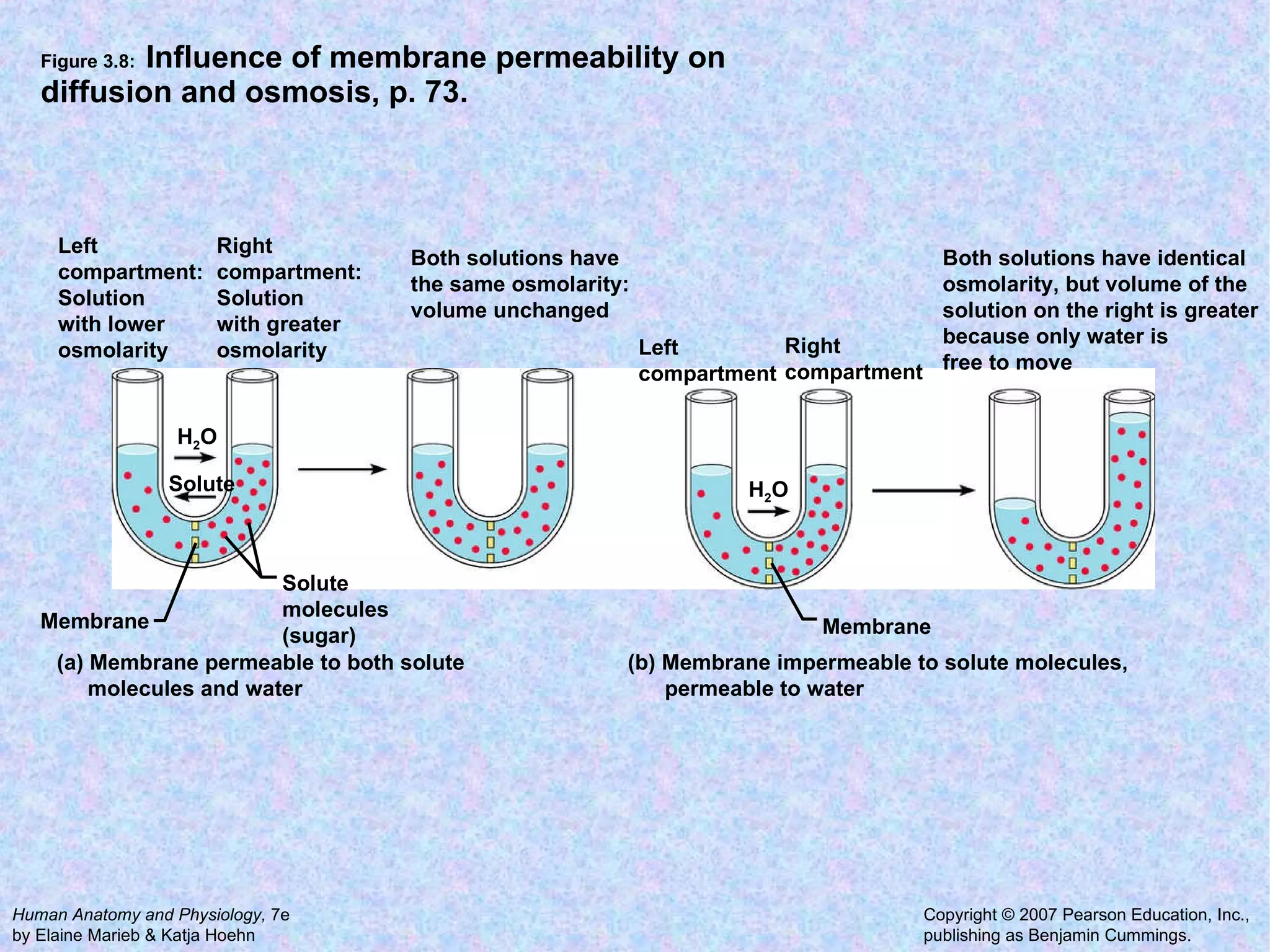
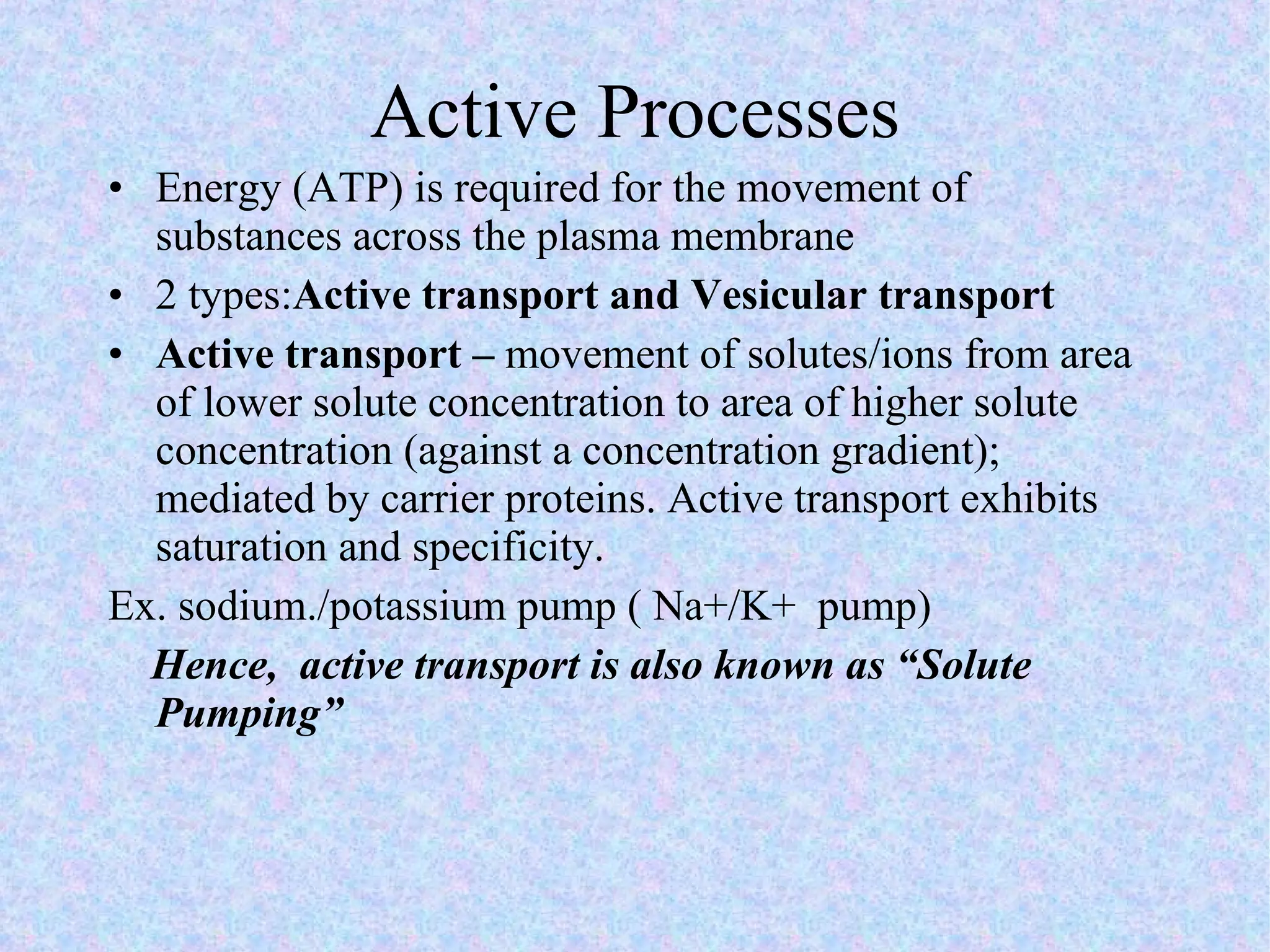
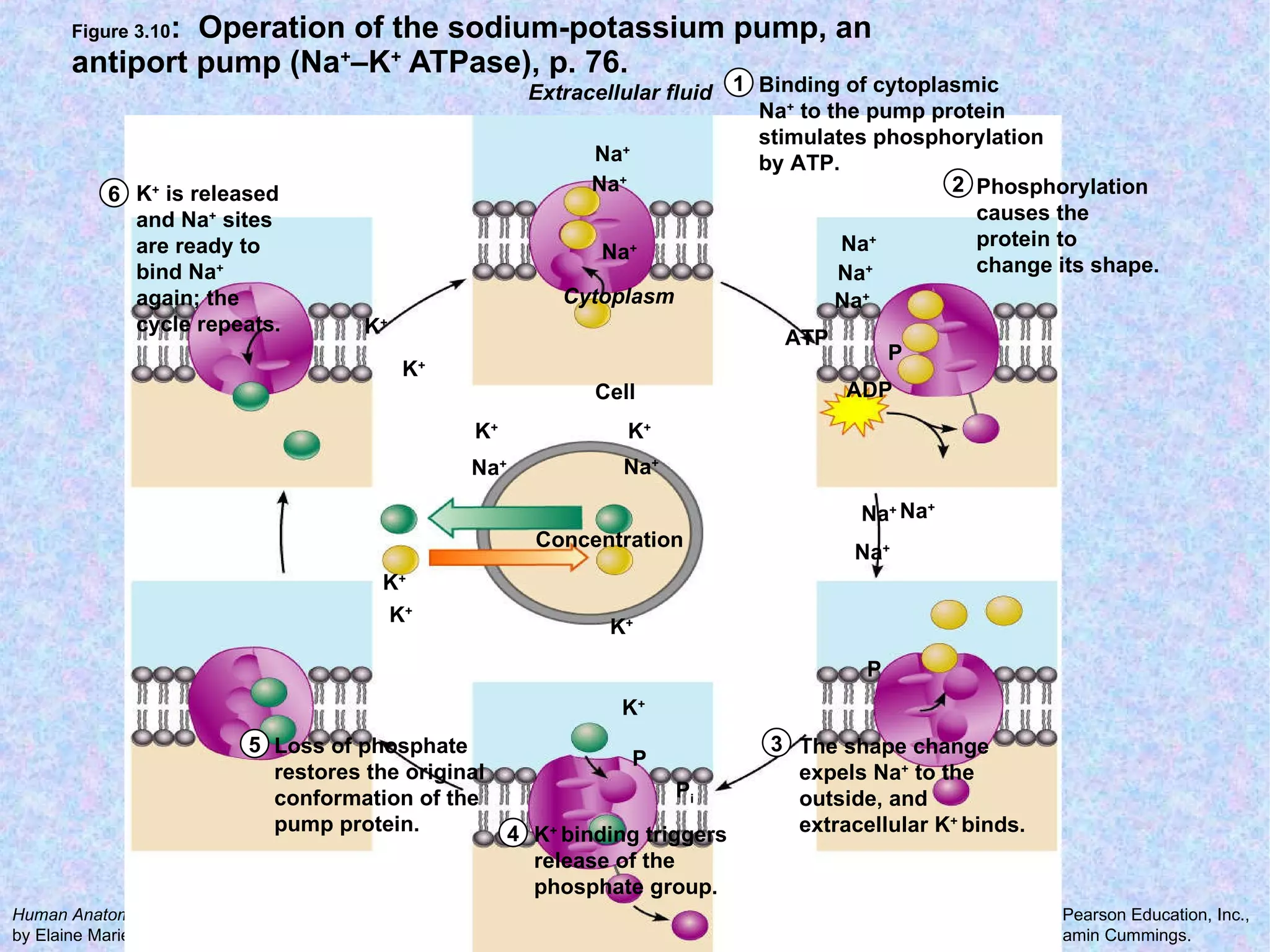
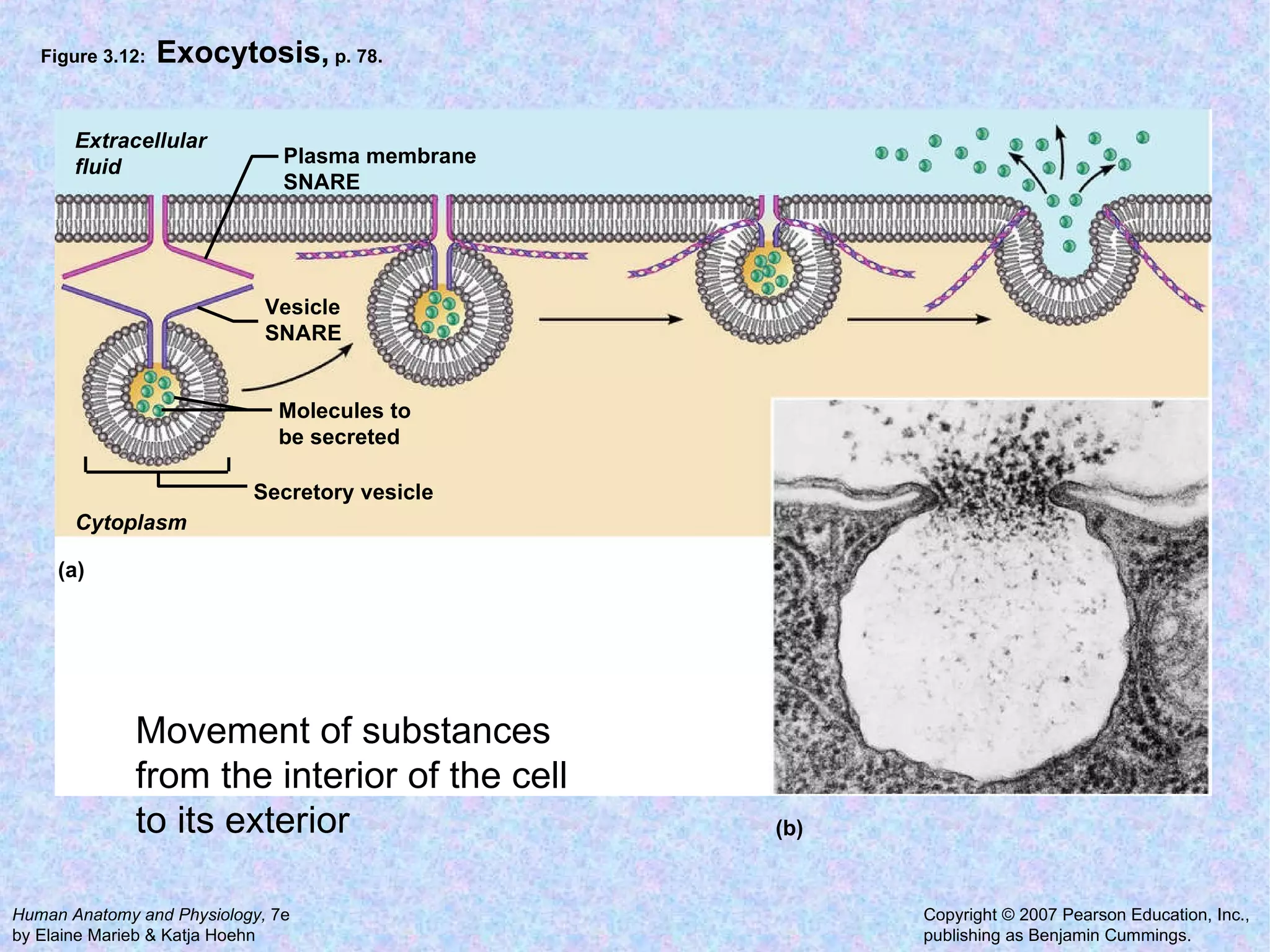
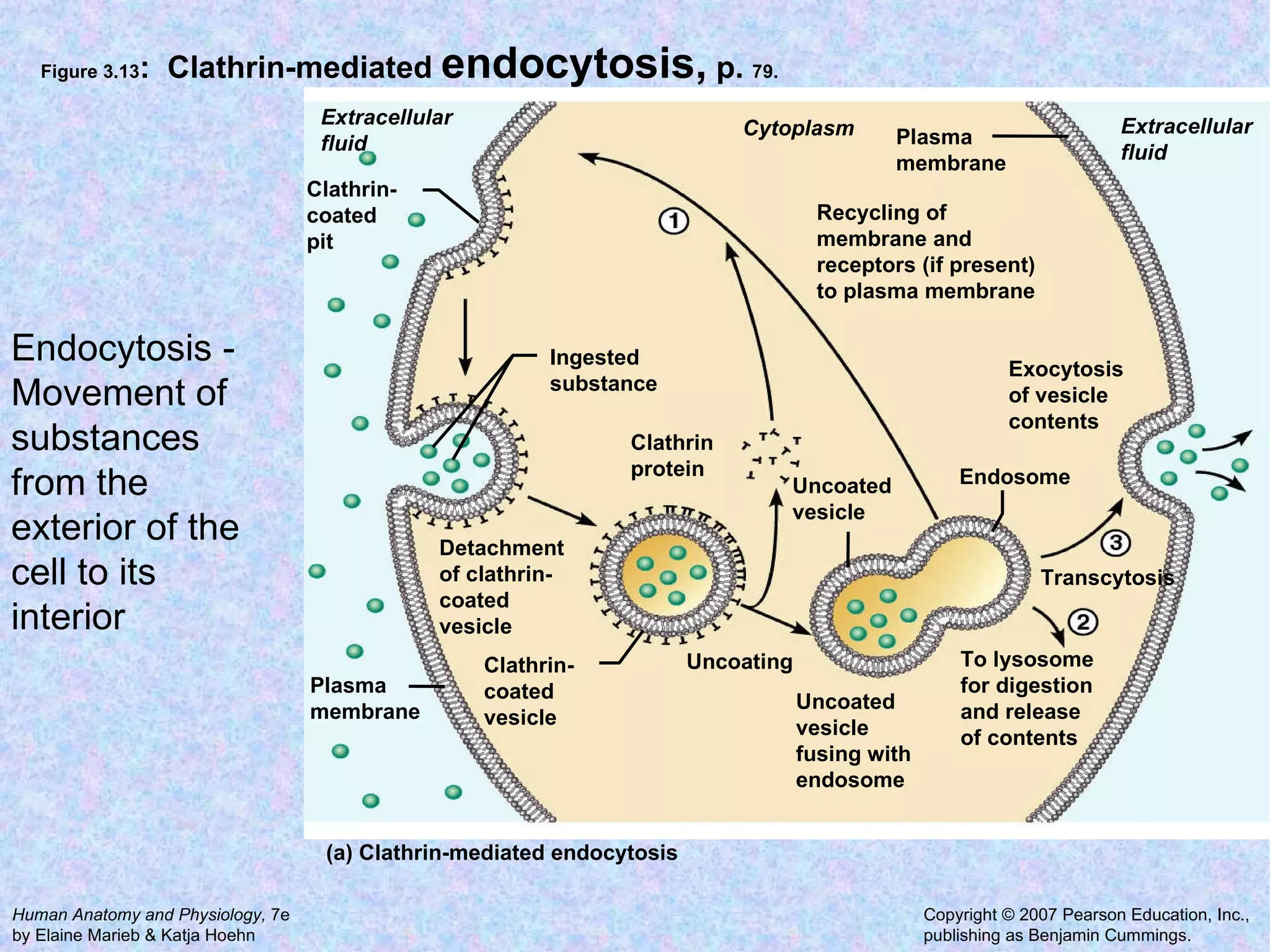
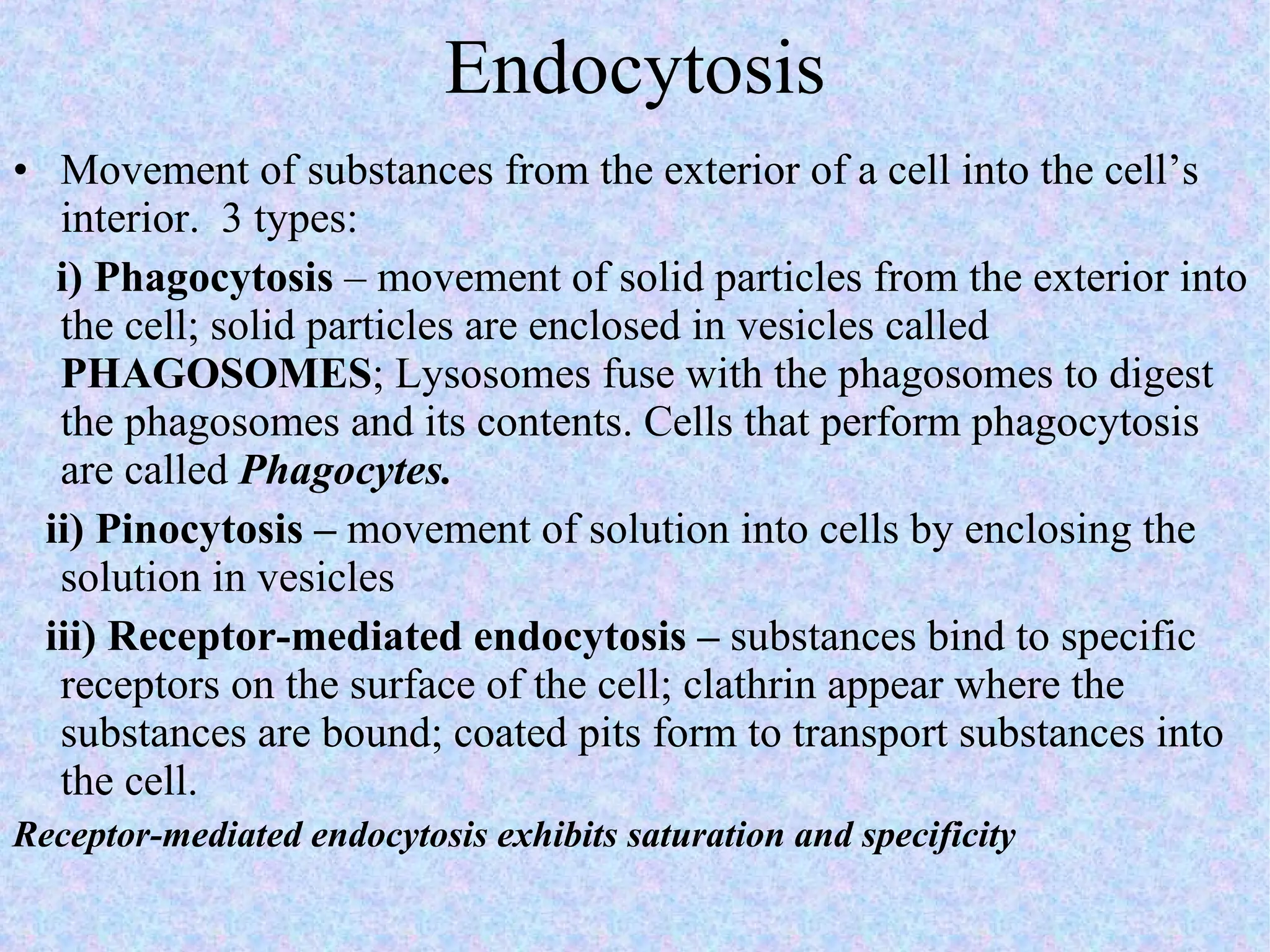
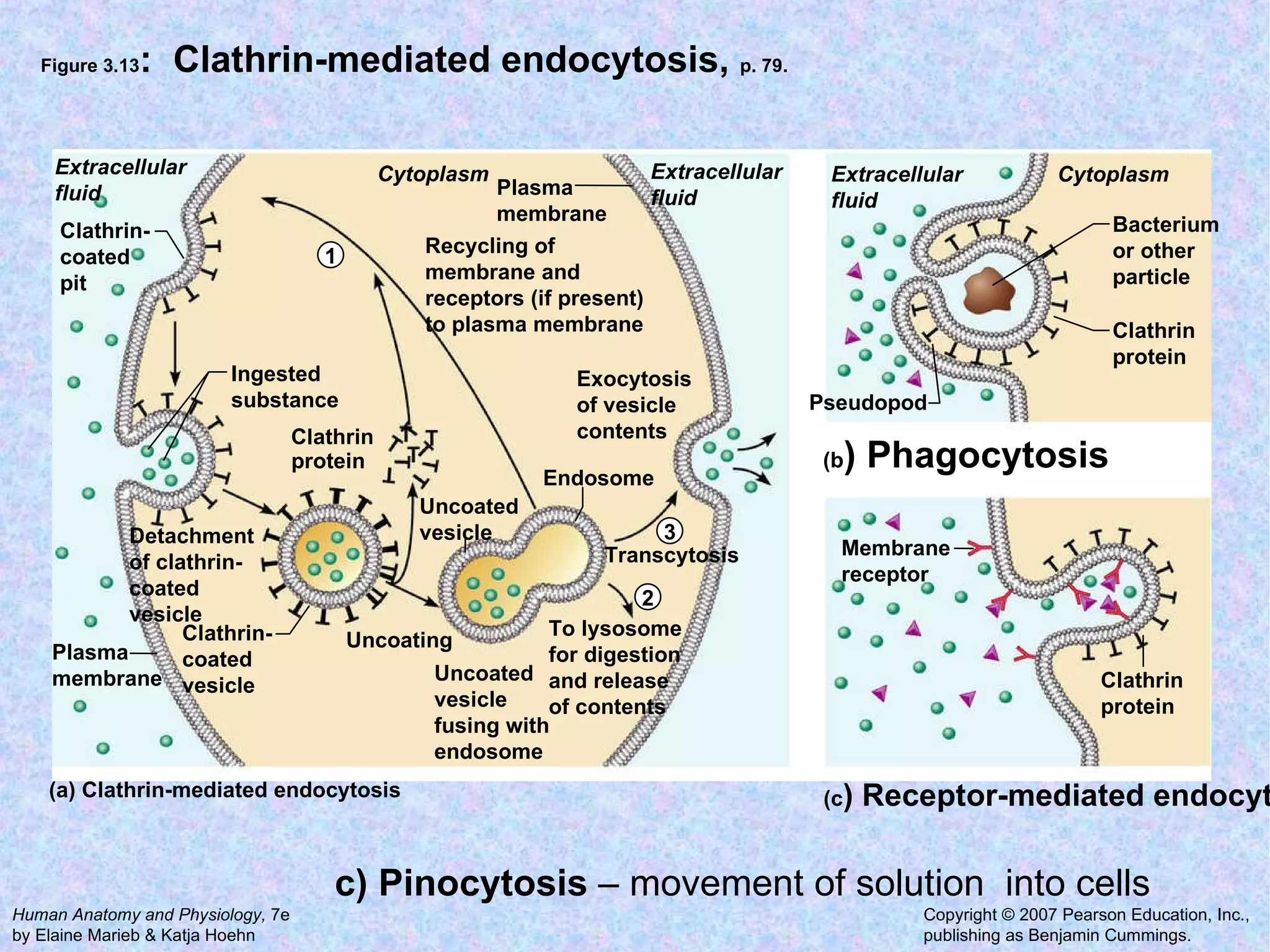
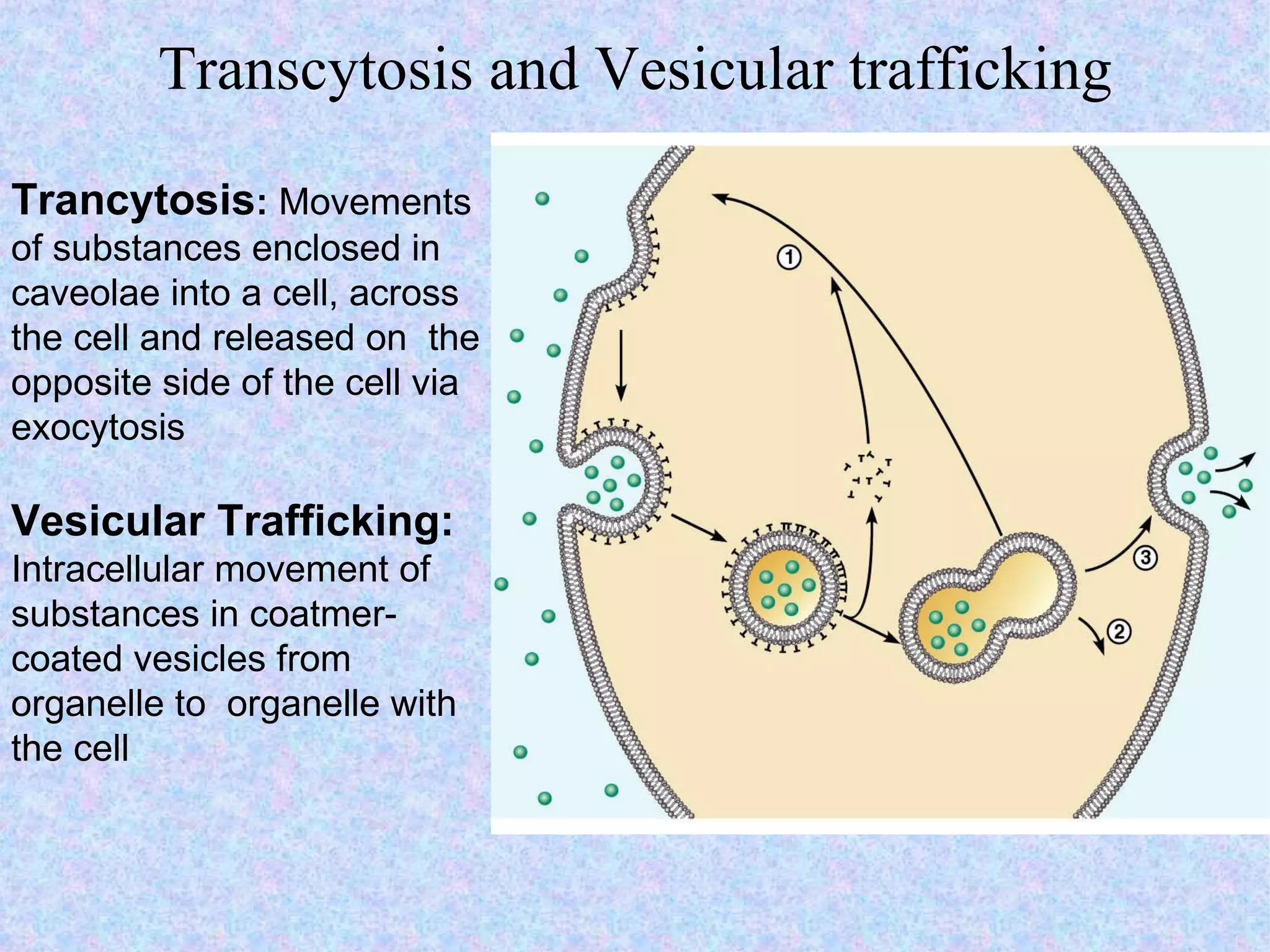
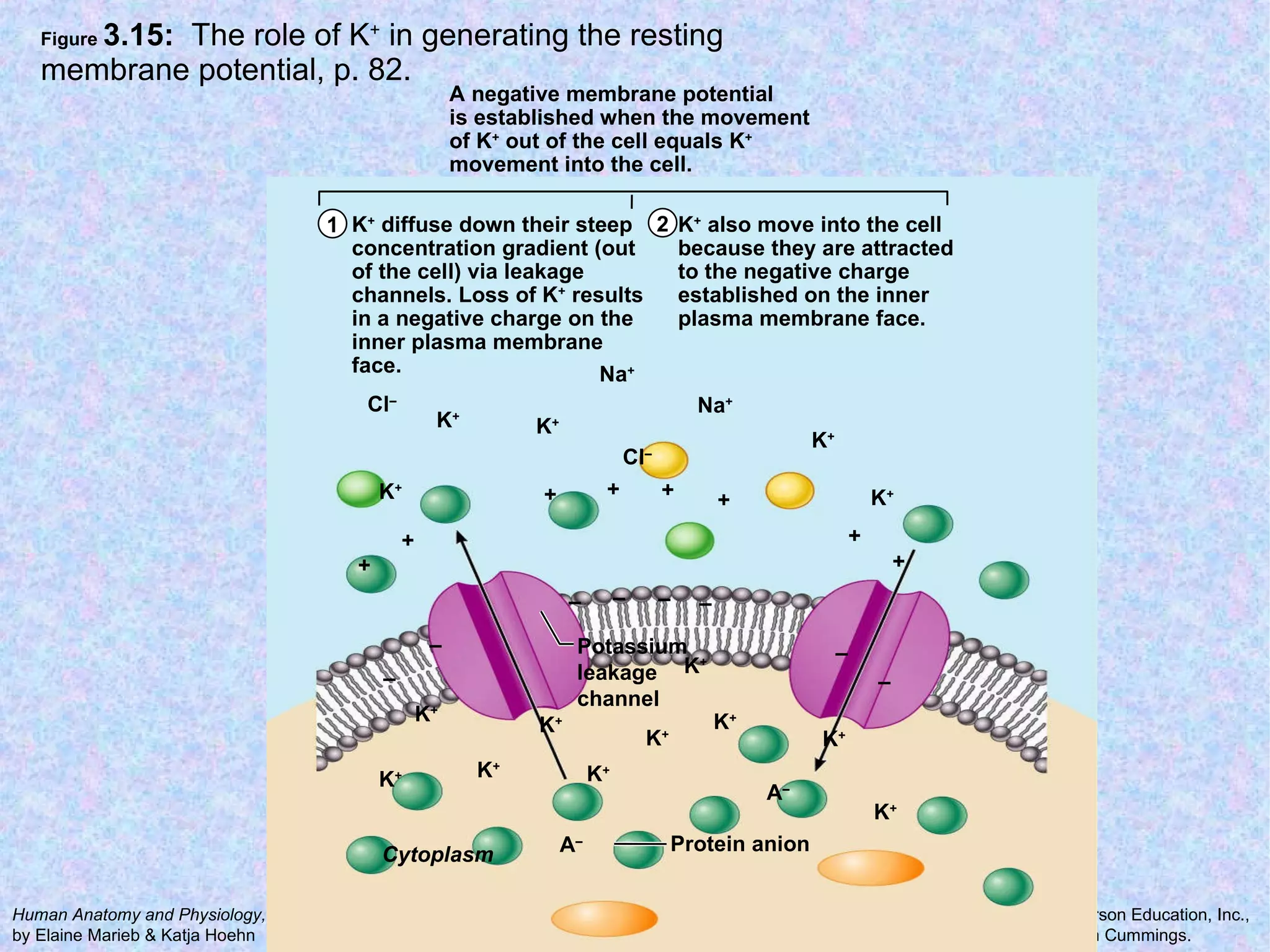
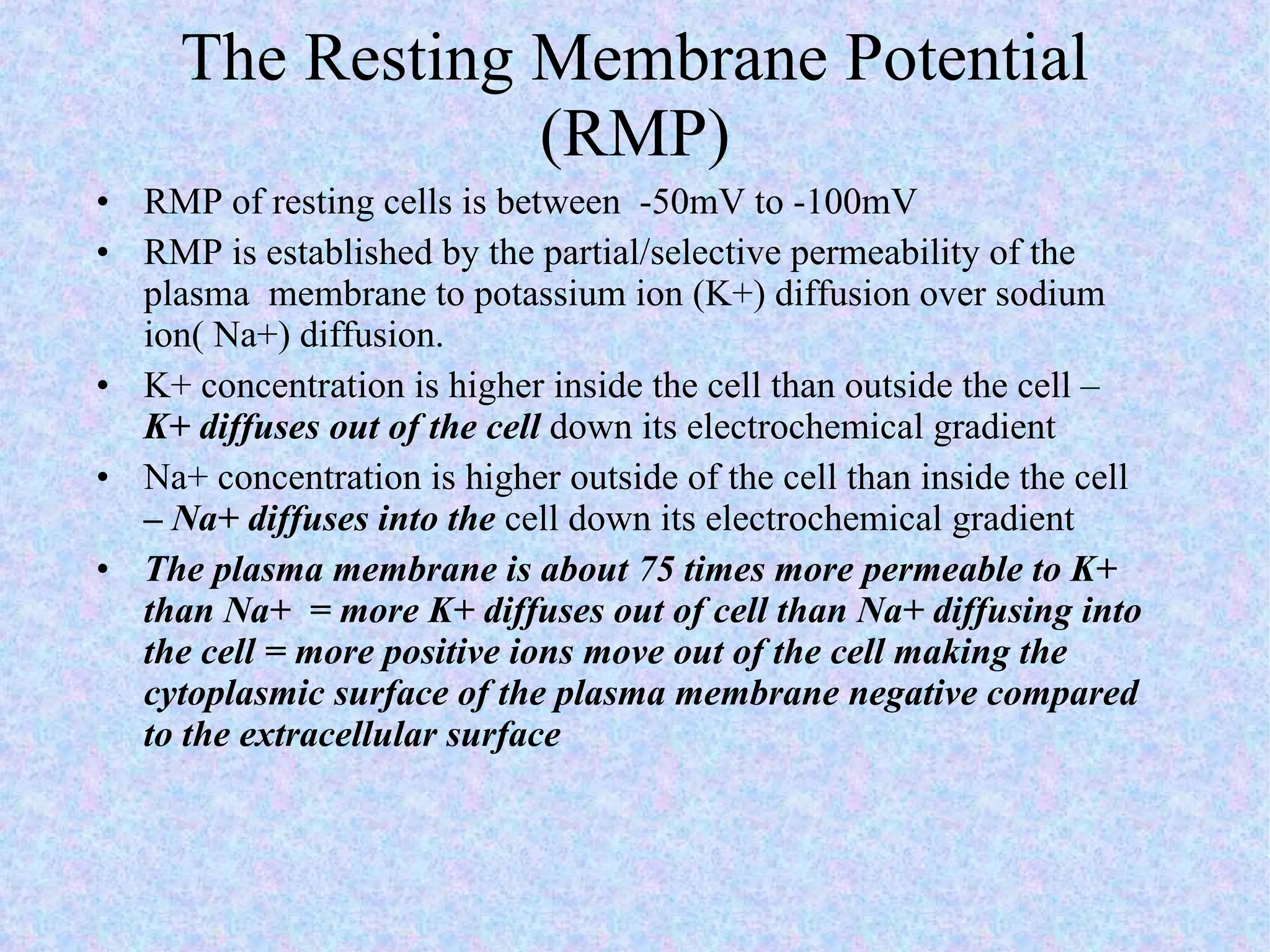
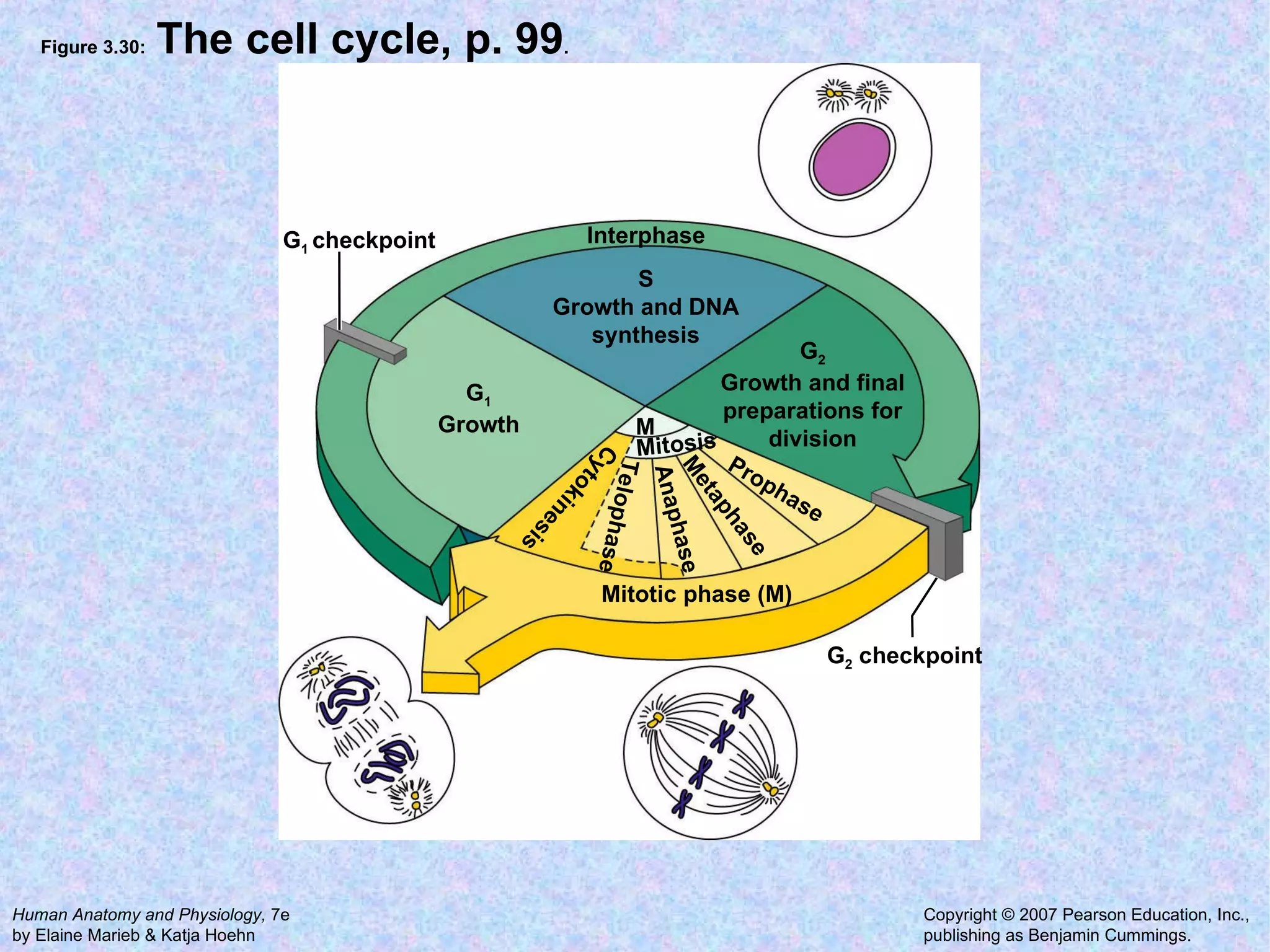
3. Transport across the plasma membrane can occur passively via diffusion and osmosis or actively via pumps that require energy. The fluid mosaic model describes the plasma membrane structure.