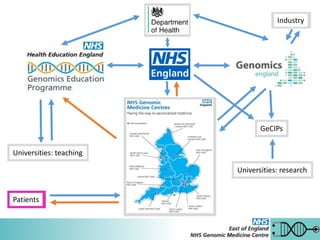
The 100,000 Genomes Project aims to sequence 100,000 genomes from 70,000 people in the UK. For rare diseases, the project seeks to provide genetic diagnoses for patients by comprehensively analyzing their full genome. It also aims to kickstart genomic medicine in the NHS and a UK genomics industry. The East of England Genomic Medicine Centre is one of several centers that recruits participants and returns results as part of this large national project.



![Genomics for Rare Disease: A Brief History
• 2010: First research showing exome sequencing could be used to identify new
rare disease genes
• 2010: Lupski sequences his genome to identify molecular basis of his CMT
• March 2011: First paper published using exome sequencing to make a diagnosis,
Nicholas Volkner
• April 2011: Launch of Deciphering Developmental Disorders (DDD) Study in UK
• Sept 2011: Clinical exome sequencing test available in US (Ambry Genetics)
• 2012: ACMG guidelines for exome/genome sequencing
• December 2012: 100,000 Genomes Project announced
• 2013: ACMG guidelines for [incidental] secondary findings
• July 2013: Rare Disease focus for 100,000 Genomes Project announced
• 2015: ~12 US labs offering clinical exome sequencing for rare disease diagnosis
• >>10,000 patients sequenced, considered standard of care in US](https://image.slidesharecdn.com/gemmachandratillakecrdncrdnsummit2017-200227181928/85/Gemma-chandratillake-crdn-crdn-summit-2017-5-320.jpg)