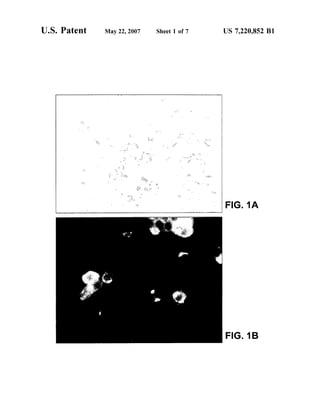
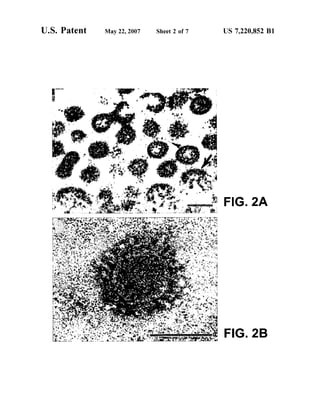
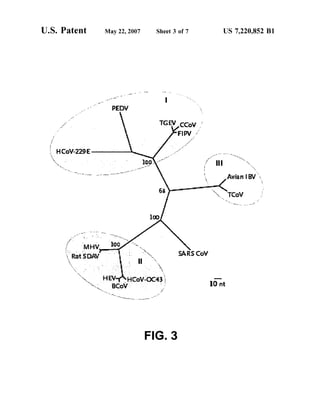
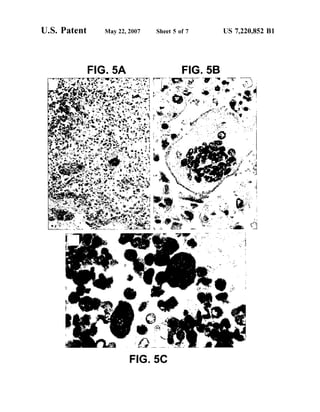
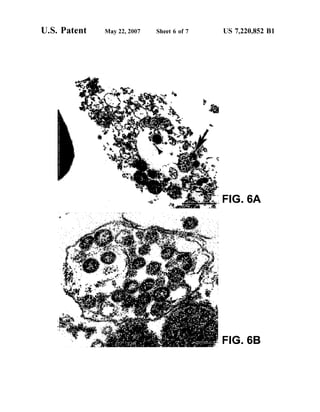
This patent document describes the isolation and characterization of a novel human coronavirus (SARS-CoV) that is the causative agent of severe acute respiratory syndrome (SARS). It provides the nucleic acid sequence of the SARS-CoV genome and the amino acid sequences of its open reading frames. Methods are described for using these molecules to detect SARS-CoV and detect infections. Immune stimulatory compositions are also provided, along with methods for their use.