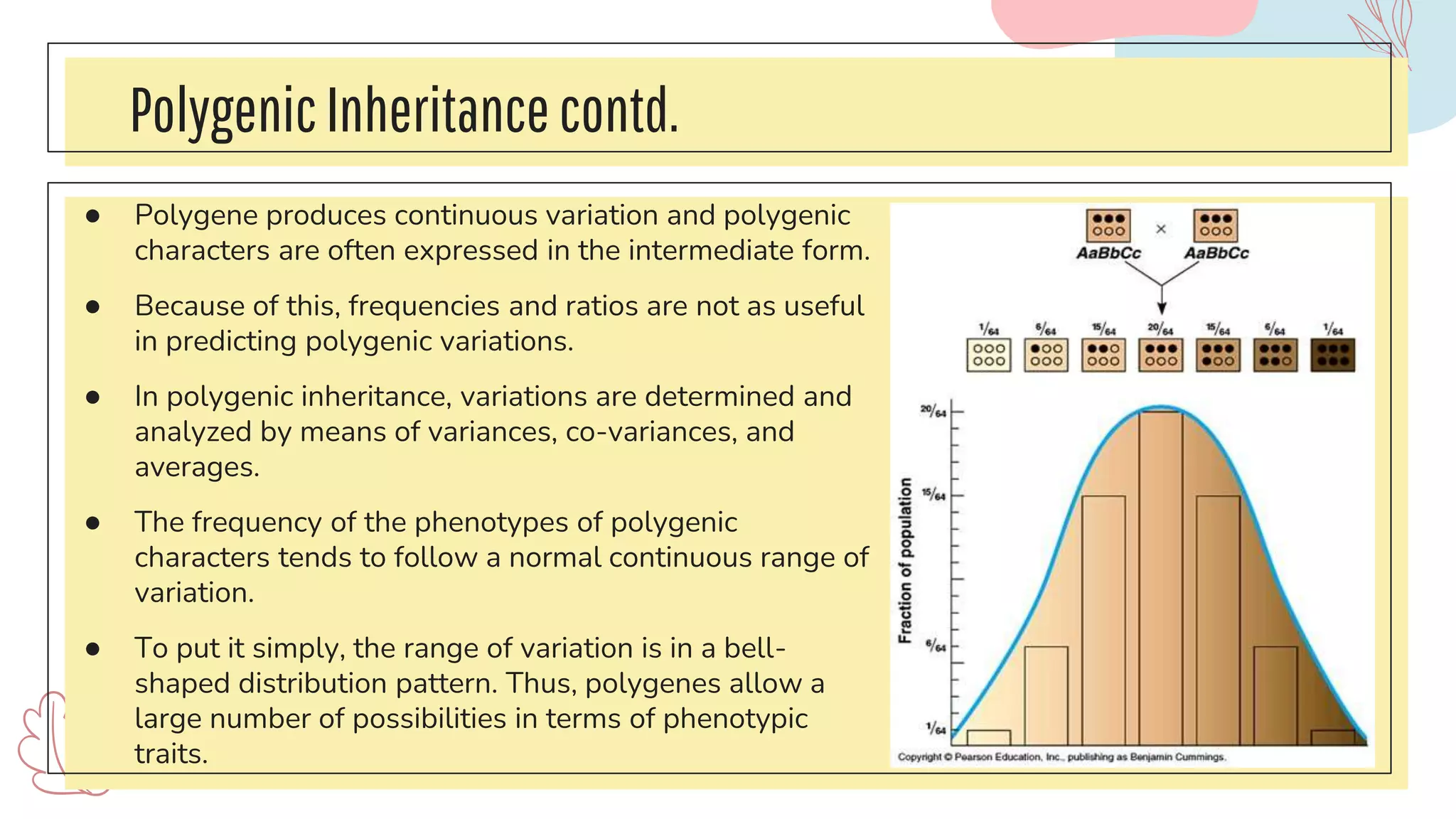
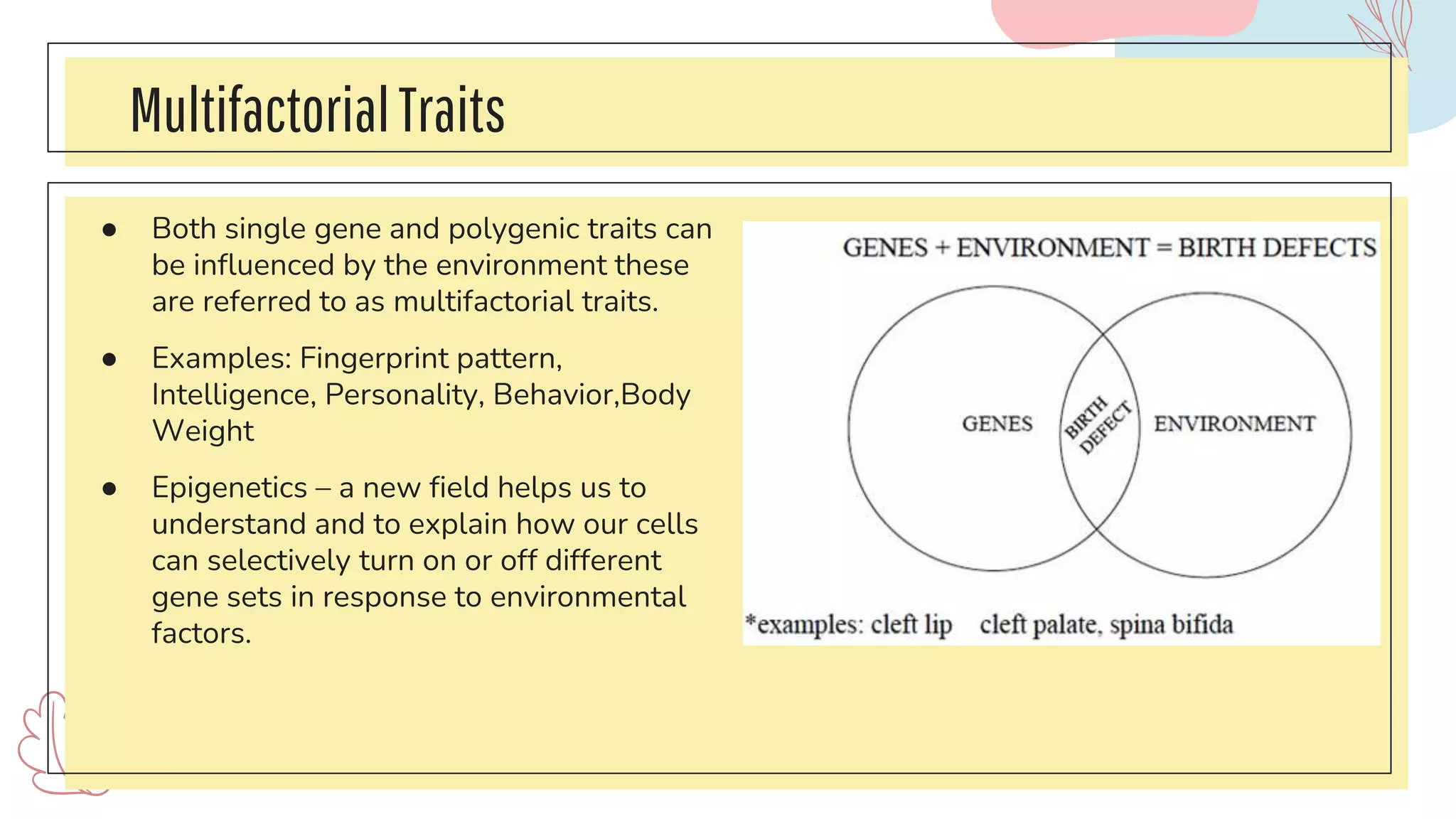
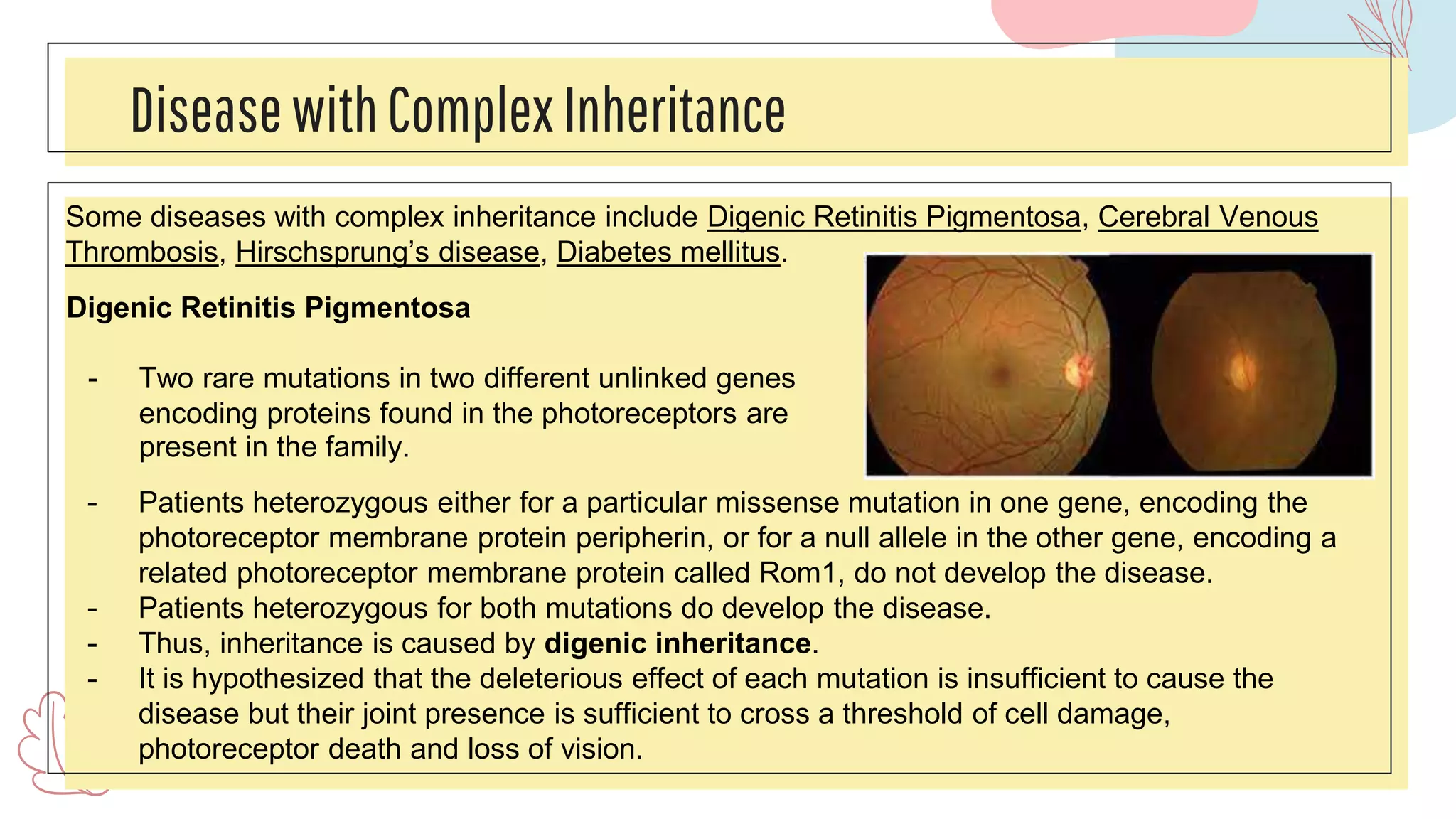
The document discusses complex traits in human genetics that do not follow Mendelian inheritance, emphasizing polygenic traits controlled by multiple genes and their interactions with environmental factors. It explains the liability/threshold model for multifactorial traits, detailing the role of genetic and environmental influences in diseases like obesity and neural tube defects. Additionally, it explores methodologies for genetic analysis and mapping of complex traits, with examples of specific diseases that exhibit complex inheritance patterns.