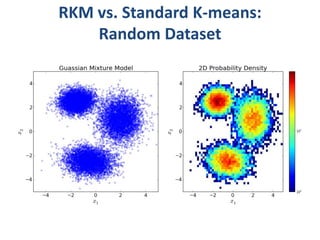
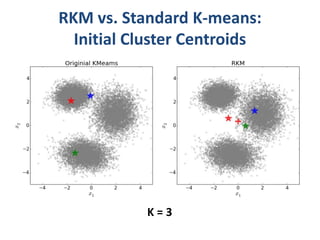
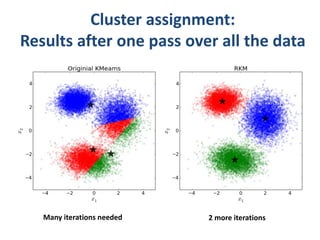
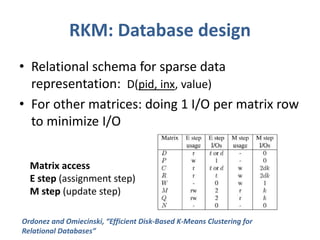
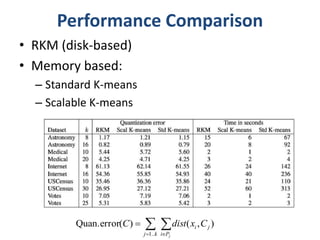
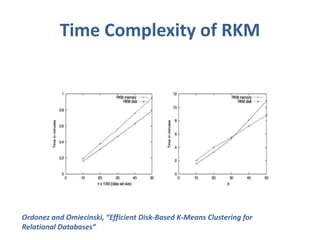
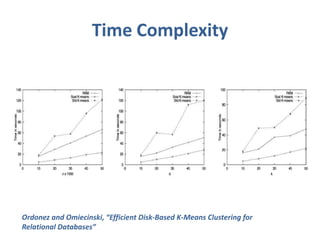
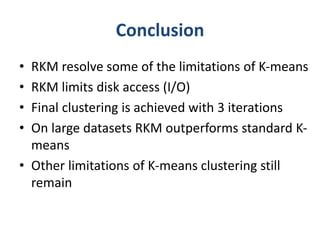
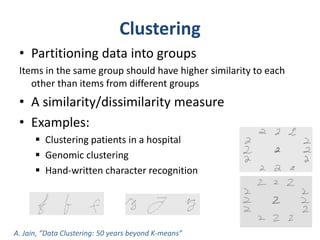
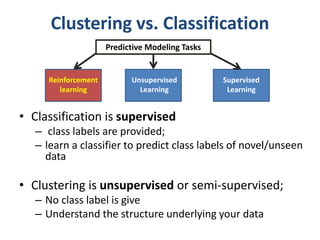
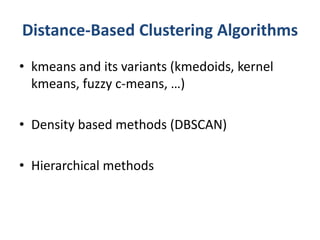
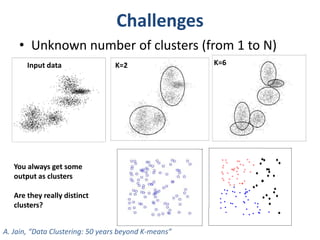
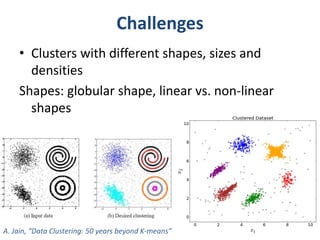
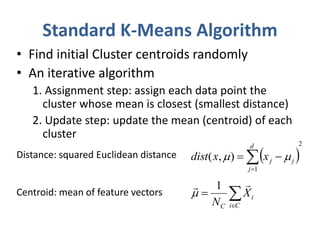
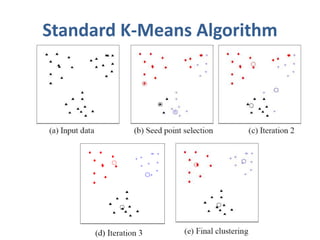
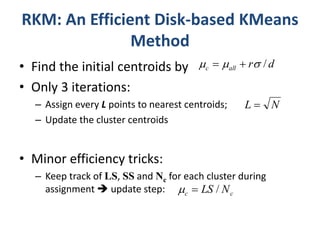
This document discusses clustering algorithms for large datasets that do not fit into main memory. It introduces the Relational K-Means (RKM) algorithm, which limits disk I/O by assigning data points in batches and updating cluster centroids after only 3 iterations. RKM stores cluster assignment and centroid data in matrices on disk and minimizes I/O by accessing matrix rows sequentially. An evaluation shows RKM outperforms standard K-means on large datasets due to its ability to handle data that does not fit in memory through efficient disk access. However, RKM does not address all limitations of K-means clustering.



![Implementation of RKM:
storing data matrices
• D input dataset
• Pj
cluster j (for j in [1..k])
• Mj, Qj, Nj
Linear Sum, Squared Sum, cluster
size
• Cj, Rj, Wj
Centroids, Variances, Weights
(accessed during update step)
C
M /
N
j j j
R Q / N
M M /
N
j j l
l k
j
t
j j j j j
W N N
1..
2
/](https://image.slidesharecdn.com/clusteringondatabasesystems-rkm-141210212816-conversion-gate02/85/Clustering-on-database-systems-rkm-12-320.jpg)