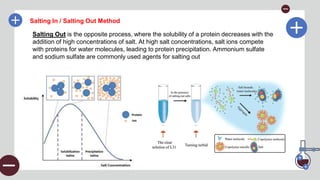
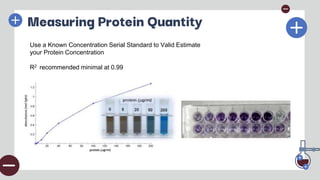
The document outlines the fundamental techniques for protein extraction and characterization in proteomics, emphasizing the importance of effective sample preparation and compatible analytical methods. It details various techniques such as RIPA buffer usage, centrifugation, salting in/out methods, dialysis, and various assays like BCA and Bradford for measuring protein concentration. Additionally, electrophoresis techniques like SDS-PAGE are discussed for the separation and characterization of proteins based on their size and charge.