Recommended
PPTX
PPT
Basics of association_mapping
PPTX
Association mapping approaches for tagging quality traits in maize
PPTX
PPTX
PPTX
Status and prospects of association mapping in crop plants
PPTX
Association mapping in plants
PPT
Association mapping for improvement of agronomic traits in rice
PPTX
PPT
Associationplants.ppt@1# Abhishek Kumar Verma
PPTX
Association mapping, GWAS, Mapping, natural population mapping
PPTX
PPTX
Genome-wideassociationmappingpresentationpptx.pptxvjifvnug
PPTX
GWAS "GENOME WIDE ASSOCIATION STUDIES" A STEP AHEAD
PDF
Association mapping in crop improvement
PPTX
Genome wide Association studies.pptx
DOCX
Report- Genome wide association studies.
PPT
Synthesis of nanoparticles Physical vapor deposition.ppt
PPT
polymeric nanoparticle, micelle Nanoparticles.ppt
PPT
Changes in Chromosome Structure revised.ppt
PPT
Chromosome banding techniques/methods.ppt
PPTX
L5-L6 - Transgenic animals and its applications.pptx
PPTX
L4 - MICROBES IN BIOPHARMING and its uses.pptx
PDF
VIGS - RNA , silencing overview .pdf
PDF
Genetically engineered resistance-virus gene derived resistance.pdf
PPT
development of marker assisted Maize.ppt
PPT
biotechnological approaches in Weed management.ppt
PPTX
LEC.5- Finger printing with markers.pptx
PPTX
LEC. 6 - Genetic Diversity in Human.pptx
PPT
Enzymes in the food industry and its uses.ppt
More Related Content
PPTX
PPT
Basics of association_mapping
PPTX
Association mapping approaches for tagging quality traits in maize
PPTX
PPTX
PPTX
Status and prospects of association mapping in crop plants
PPTX
Association mapping in plants
PPT
Association mapping for improvement of agronomic traits in rice
Similar to Association Mapping concepts and types.pptx
PPTX
PPT
Associationplants.ppt@1# Abhishek Kumar Verma
PPTX
Association mapping, GWAS, Mapping, natural population mapping
PPTX
PPTX
Genome-wideassociationmappingpresentationpptx.pptxvjifvnug
PPTX
GWAS "GENOME WIDE ASSOCIATION STUDIES" A STEP AHEAD
PDF
Association mapping in crop improvement
PPTX
Genome wide Association studies.pptx
DOCX
Report- Genome wide association studies.
More from vino vidhu
PPT
Synthesis of nanoparticles Physical vapor deposition.ppt
PPT
polymeric nanoparticle, micelle Nanoparticles.ppt
PPT
Changes in Chromosome Structure revised.ppt
PPT
Chromosome banding techniques/methods.ppt
PPTX
L5-L6 - Transgenic animals and its applications.pptx
PPTX
L4 - MICROBES IN BIOPHARMING and its uses.pptx
PDF
VIGS - RNA , silencing overview .pdf
PDF
Genetically engineered resistance-virus gene derived resistance.pdf
PPT
development of marker assisted Maize.ppt
PPT
biotechnological approaches in Weed management.ppt
PPTX
LEC.5- Finger printing with markers.pptx
PPTX
LEC. 6 - Genetic Diversity in Human.pptx
PPT
Enzymes in the food industry and its uses.ppt
PPT
Food laws and regulations and its functions.ppt
PPT
association of human disc degenrative diasease.ppt
PPT
food safety, and types of hazards in food
PPT
food standards - HACCP concepts and principles
PPT
Food borne pathogens and microborganisms .ppt
PPT
non thermal preservation and thermal.ppt
PPT
Mode of Action of herbicides and classification.ppt
Recently uploaded
PPTX
Appreciations - Jan 26 01.pptxkkkmmkmkmkmkm
PDF
West Hatch High School - GCSE Media Specification
PPTX
Drug Distribution in Pharmacology: Mechanisms, Barriers, Factors & Volume of ...
PPTX
That long silence - Novel by Shashi Deshapande
PPTX
Mohan - "A man of silence and authority"
PPTX
Metabolism ( BIOCHEMISTRY & CLINICAL PATHOLOGY )
PPTX
Unit 3- Culture.pptx....................
PPTX
West Hatch High School - GCSE Spanish Presentation
PDF
Information about Presentation strategies
PPTX
The night at deoli - Ruskin bond's story of first love
PPTX
"Aristotle : Father Of Western Philosophy"
PDF
Chapter 05 Drug Acting on CNS Sedatives & Hypnotics | Antipsychotics.pdf
PPTX
Day 2 ppt english.powerpoint presentation ppt
PDF
West Hatch High School - GCSE French Specification
PPTX
HABIT. Cognitive process. I Semester Shilpa Hotakar pptx
PPTX
West Hatch High School -- GCSE Geography
PDF
Information about the author Shashi Deshpande
PPTX
Drugs modulating serotonergic system.pptx
PPTX
Master of Punjabi Short Story "kartar singh duggal
PPTX
West Hatch High School - GCSE Art Presentation
Association Mapping concepts and types.pptx 1. 2. Association Mapping
“Association analysis, also known as LD mapping or
association mapping, is a population-based survey used to
identify trait-marker relationships based on linkage
disequilibrium”
(Flint-Garcia et al. 2003)
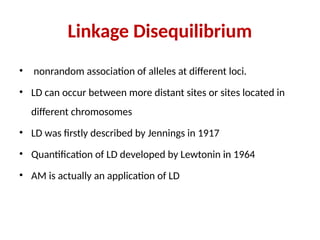
3. Linkage Disequilibrium
• nonrandom association of alleles at different loci.
• LD can occur between more distant sites or sites located in
different chromosomes
• LD was firstly described by Jennings in 1917
• Quantification of LD developed by Lewtonin in 1964
• AM is actually an application of LD
4. Pros
• AM offers three main advantages over linkage mapping:
– Mapping resolution, allele number and time saving in
establishing a marker-trait association and its application
in a breeding program (Flint-Garcia et al., 2003).
• availability of broader genetic variations with wider
background for marker-trait correlations
• no need for the development of expensive and tedious
biparental populations
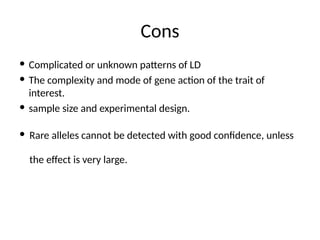
5. Cons
Complicated or unknown patterns of LD
The complexity and mode of gene action of the trait of
interest.
sample size and experimental design.
Rare alleles cannot be detected with good confidence, unless
the effect is very large.