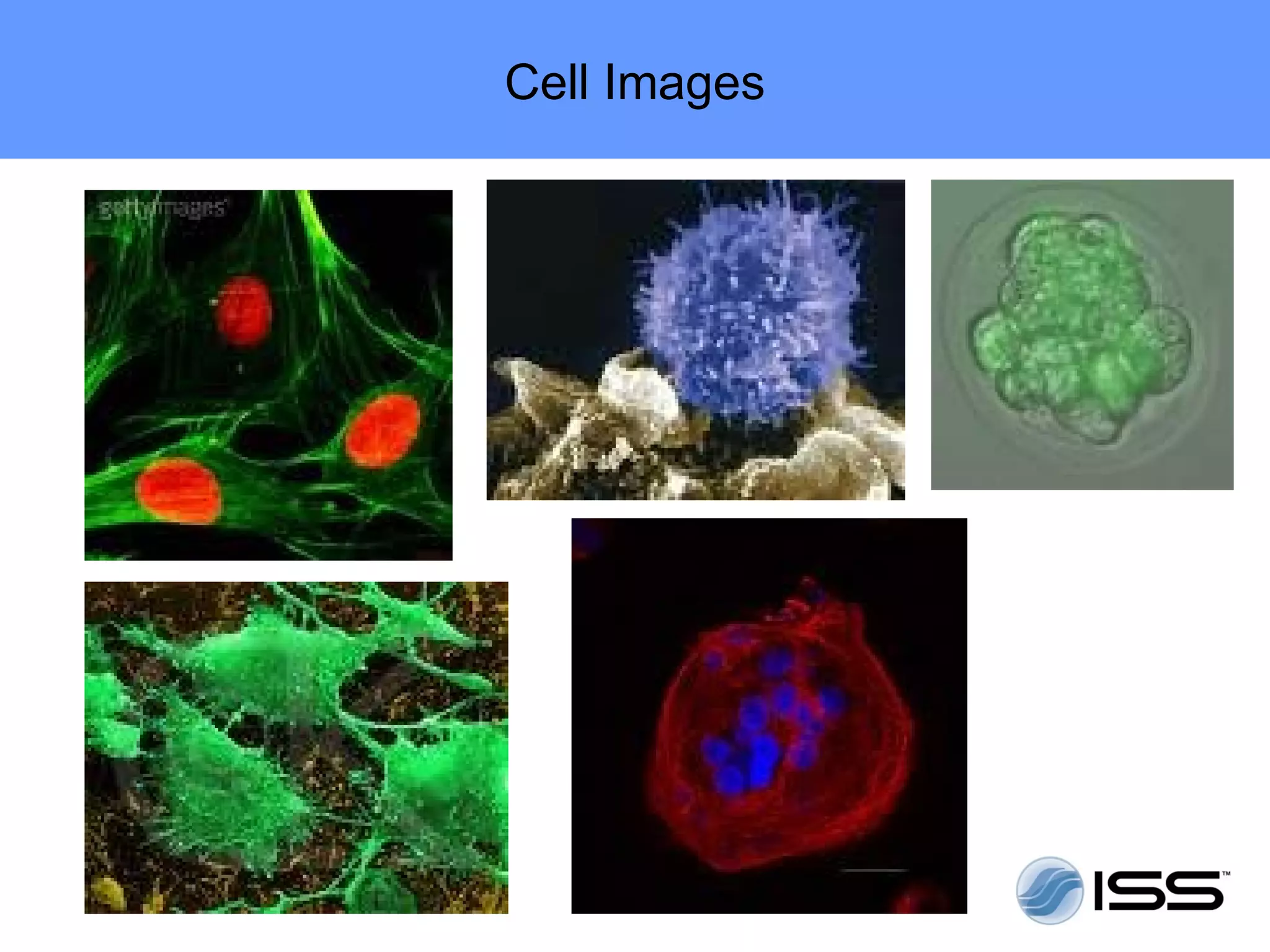
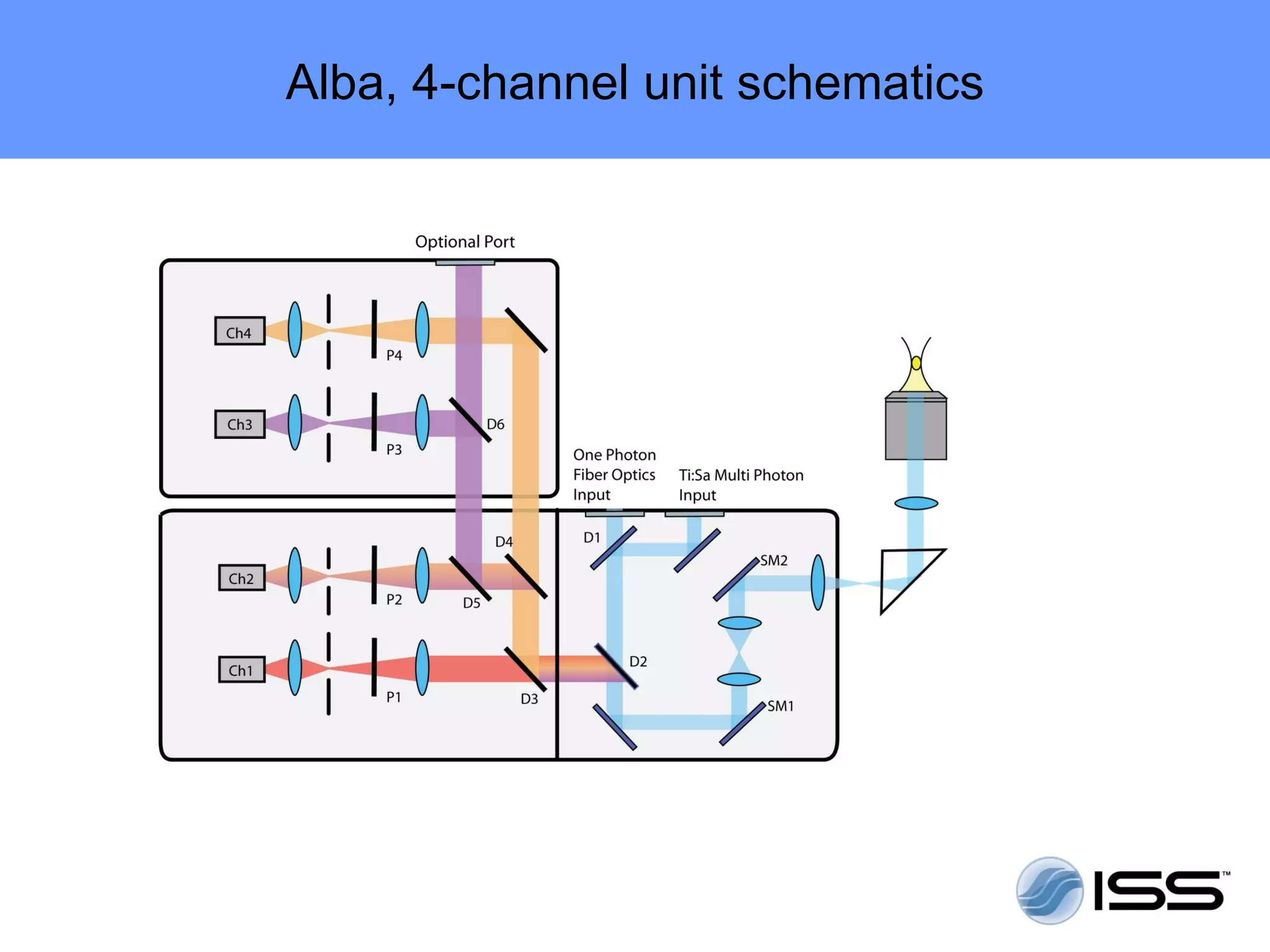
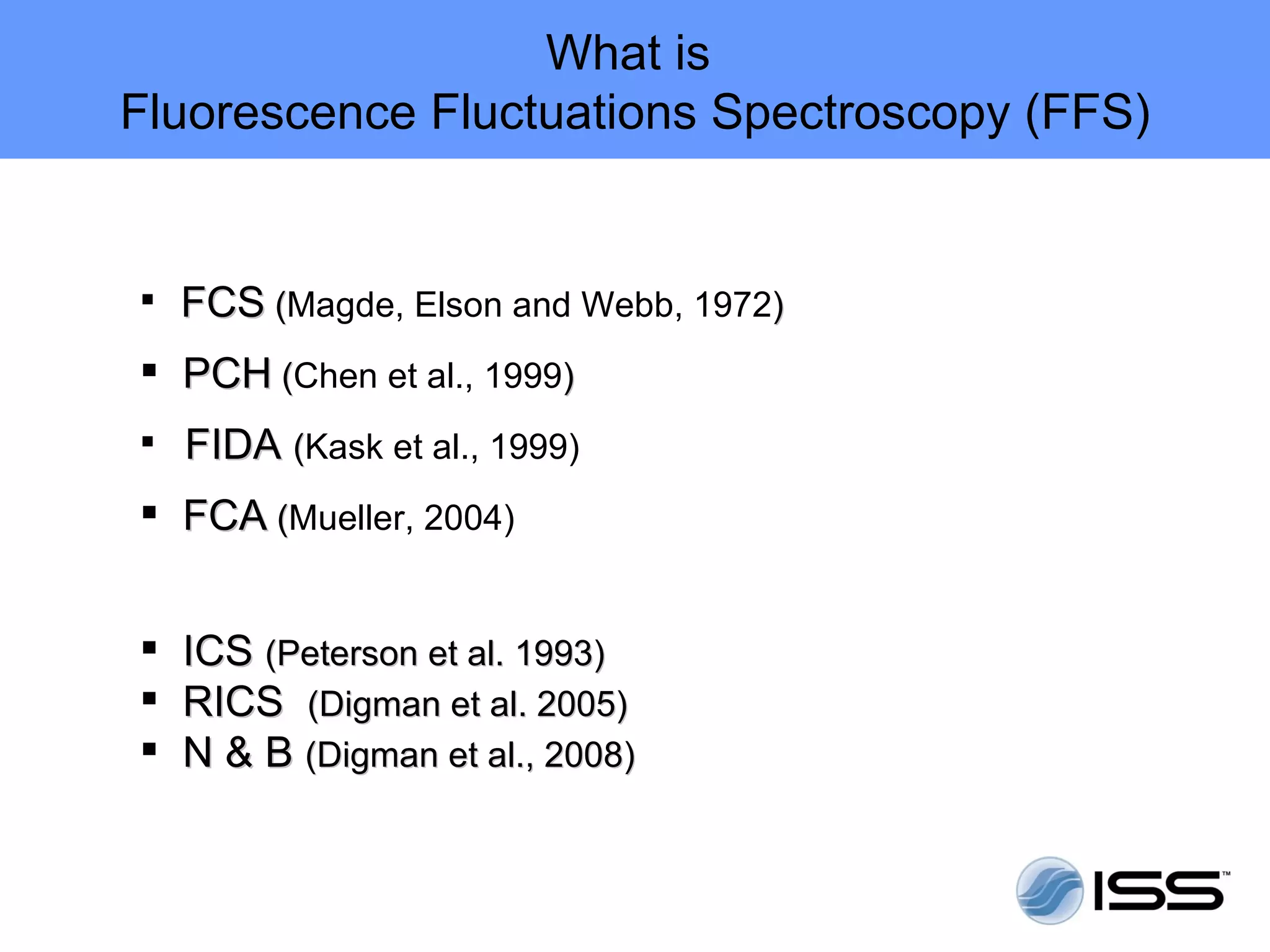
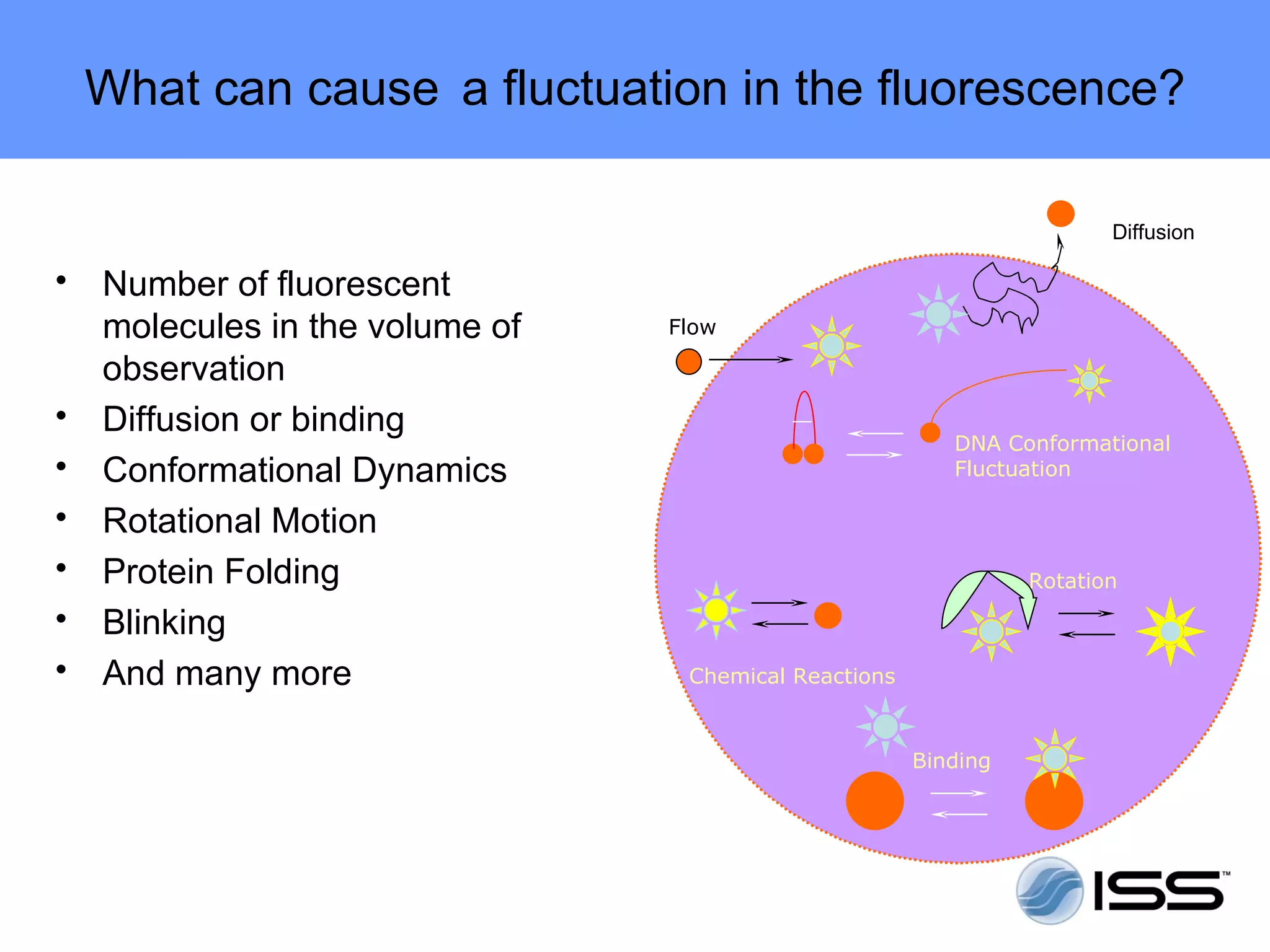
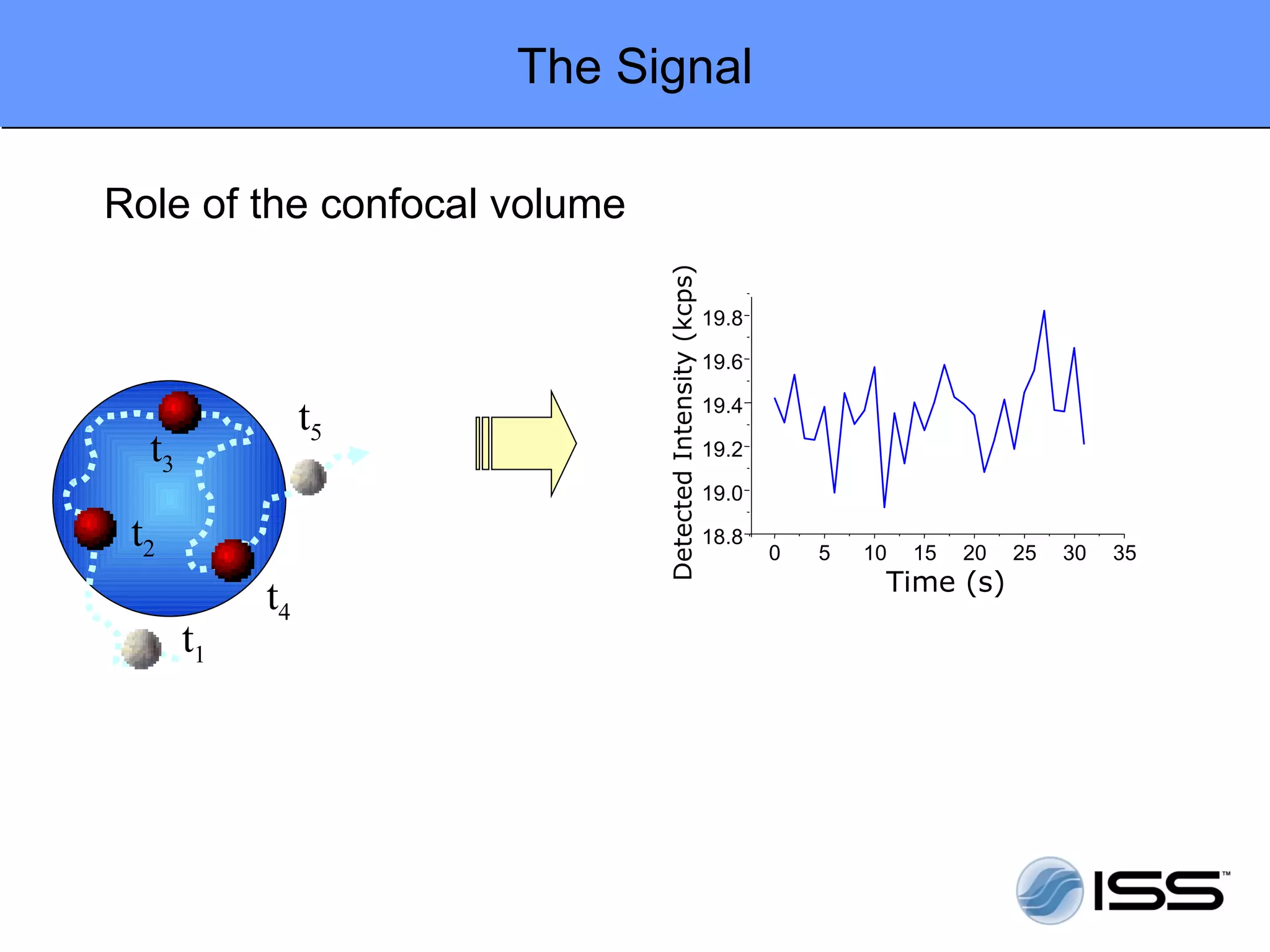
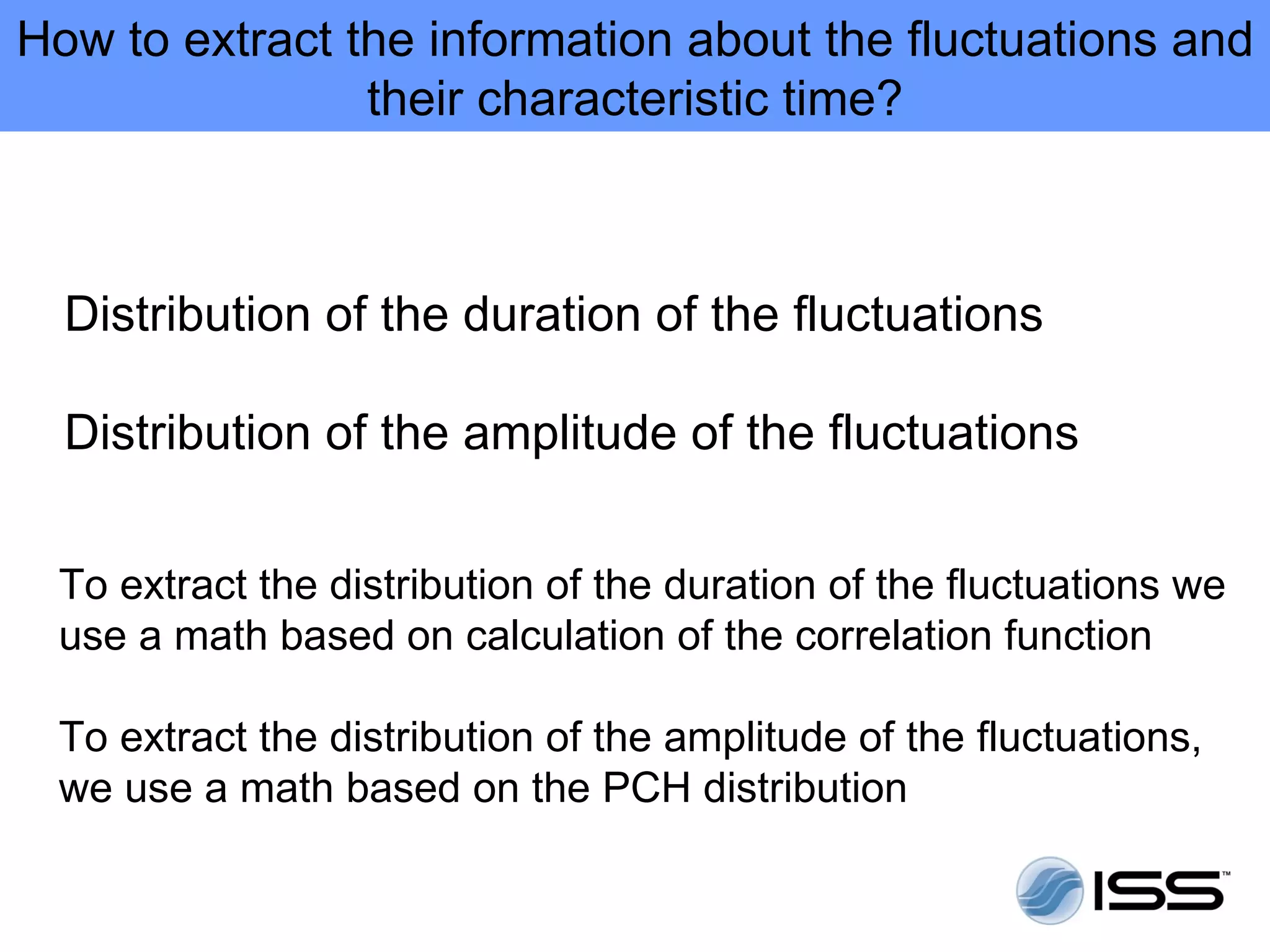
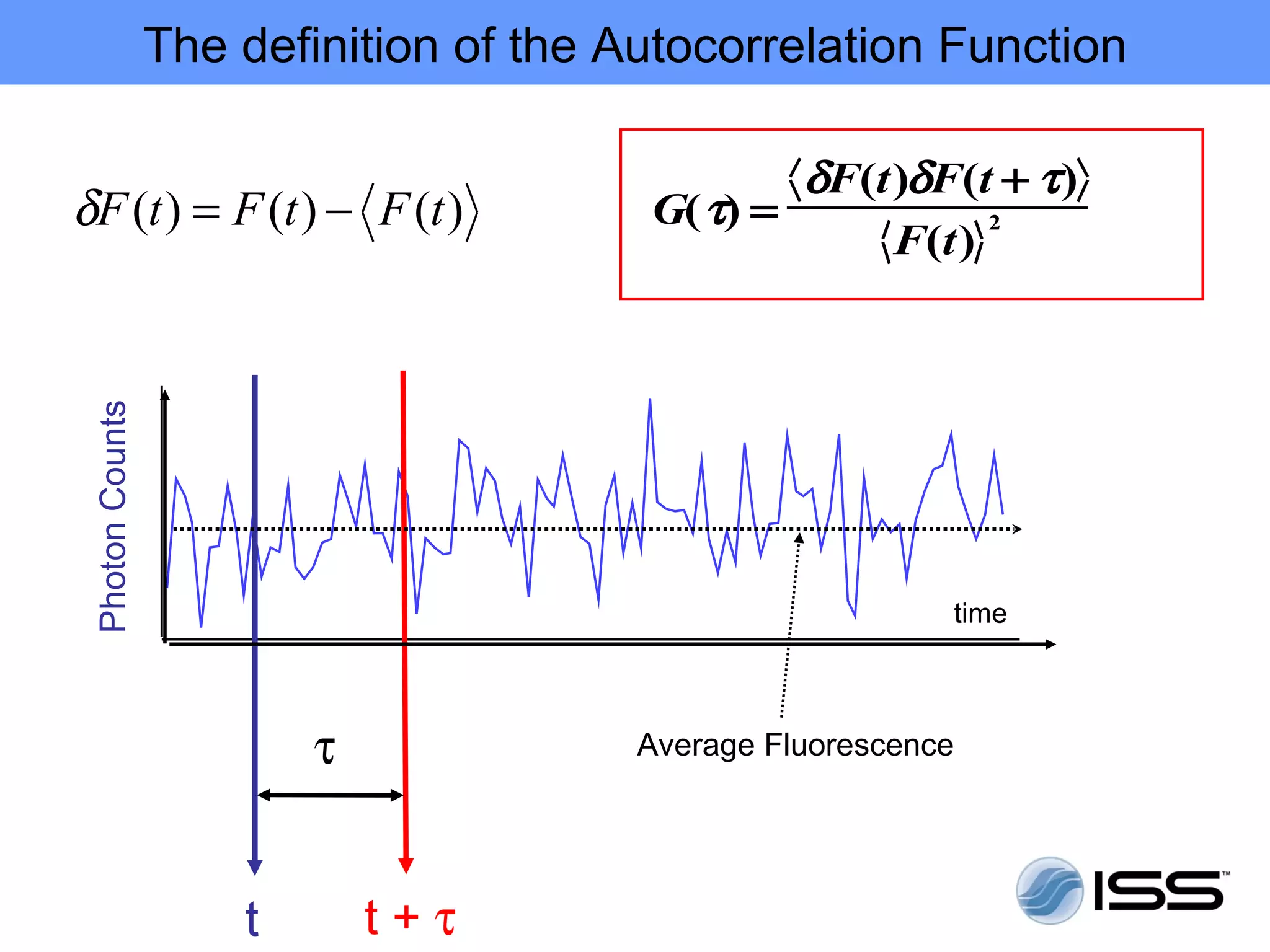
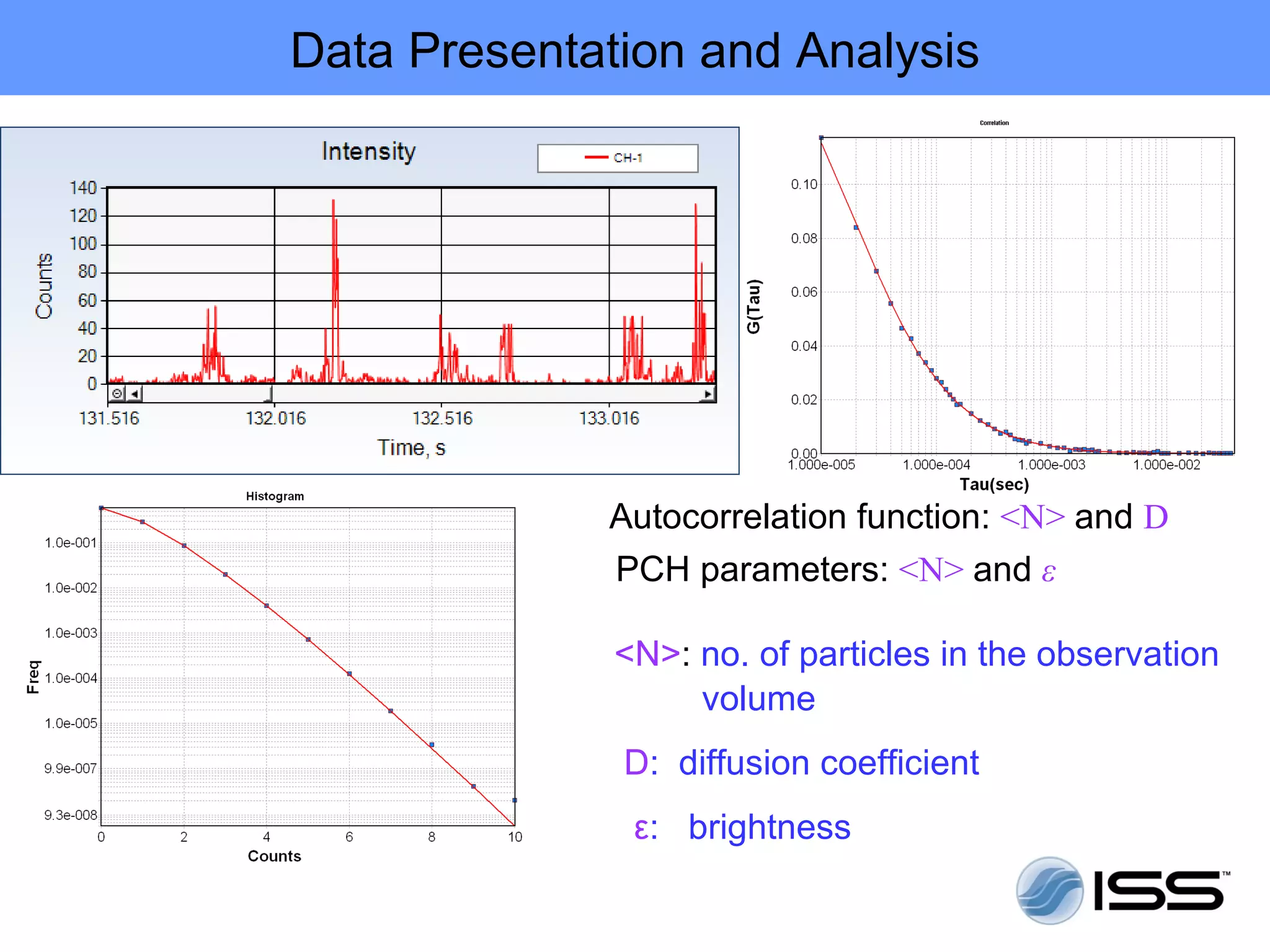
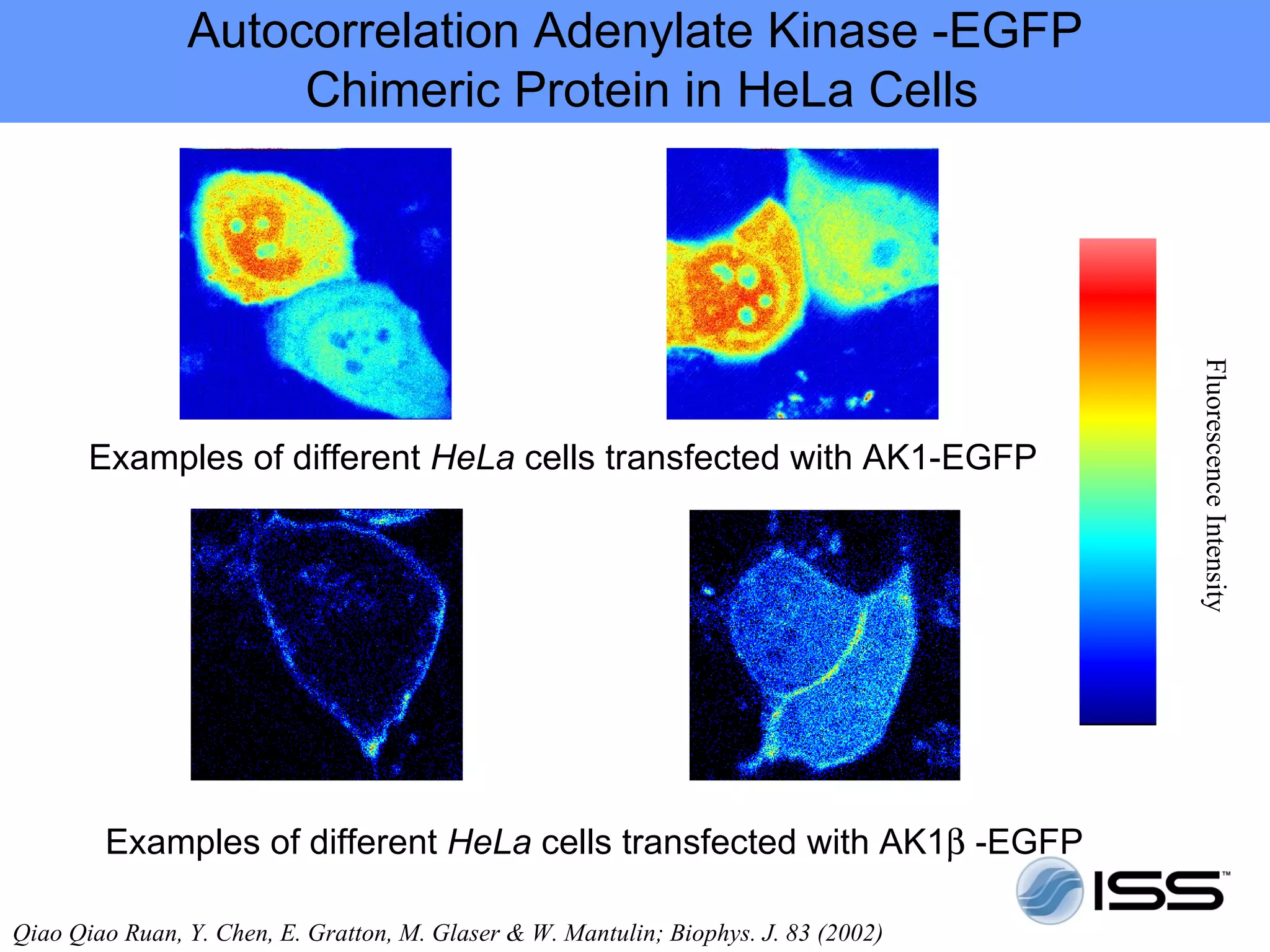
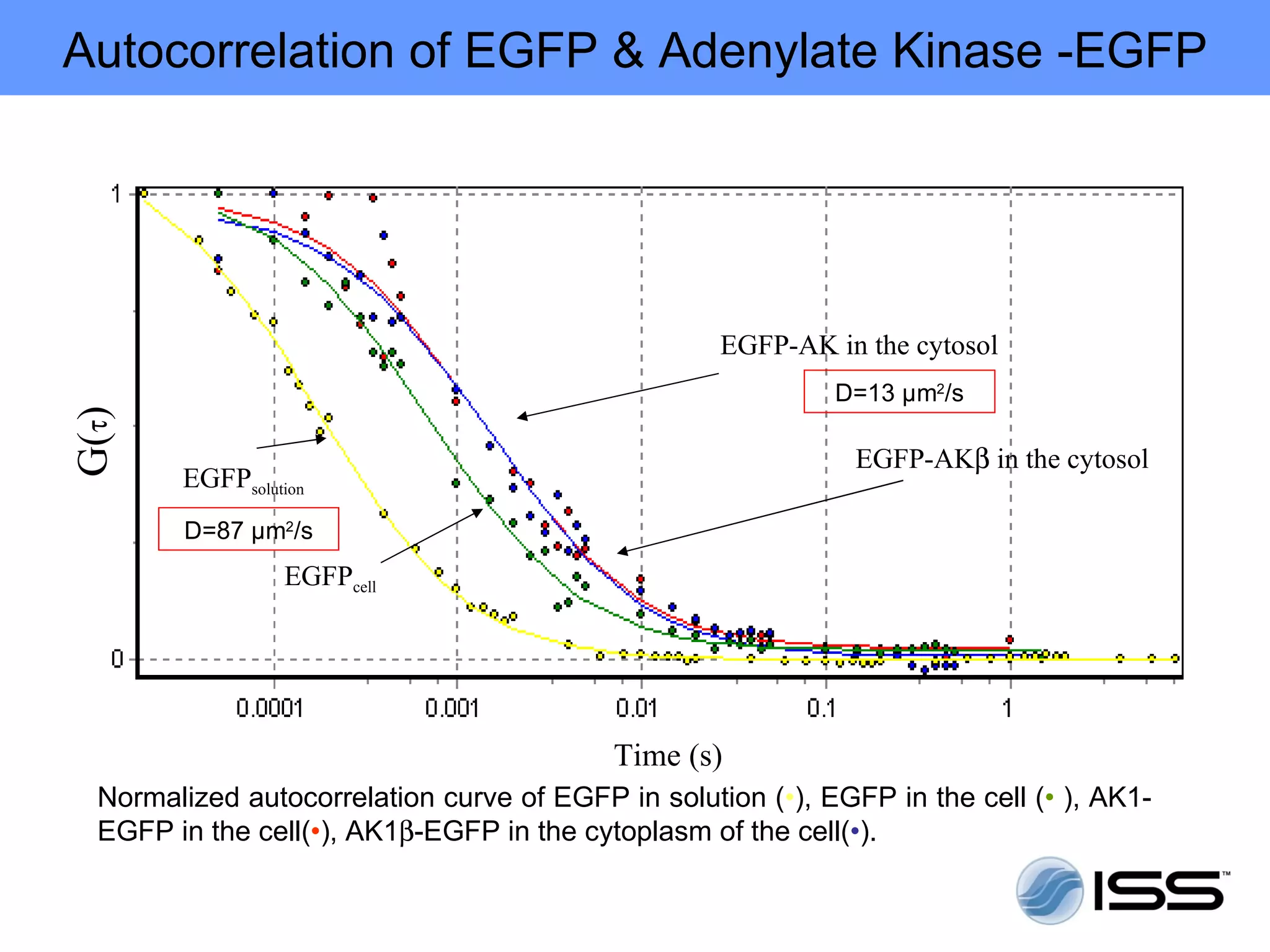
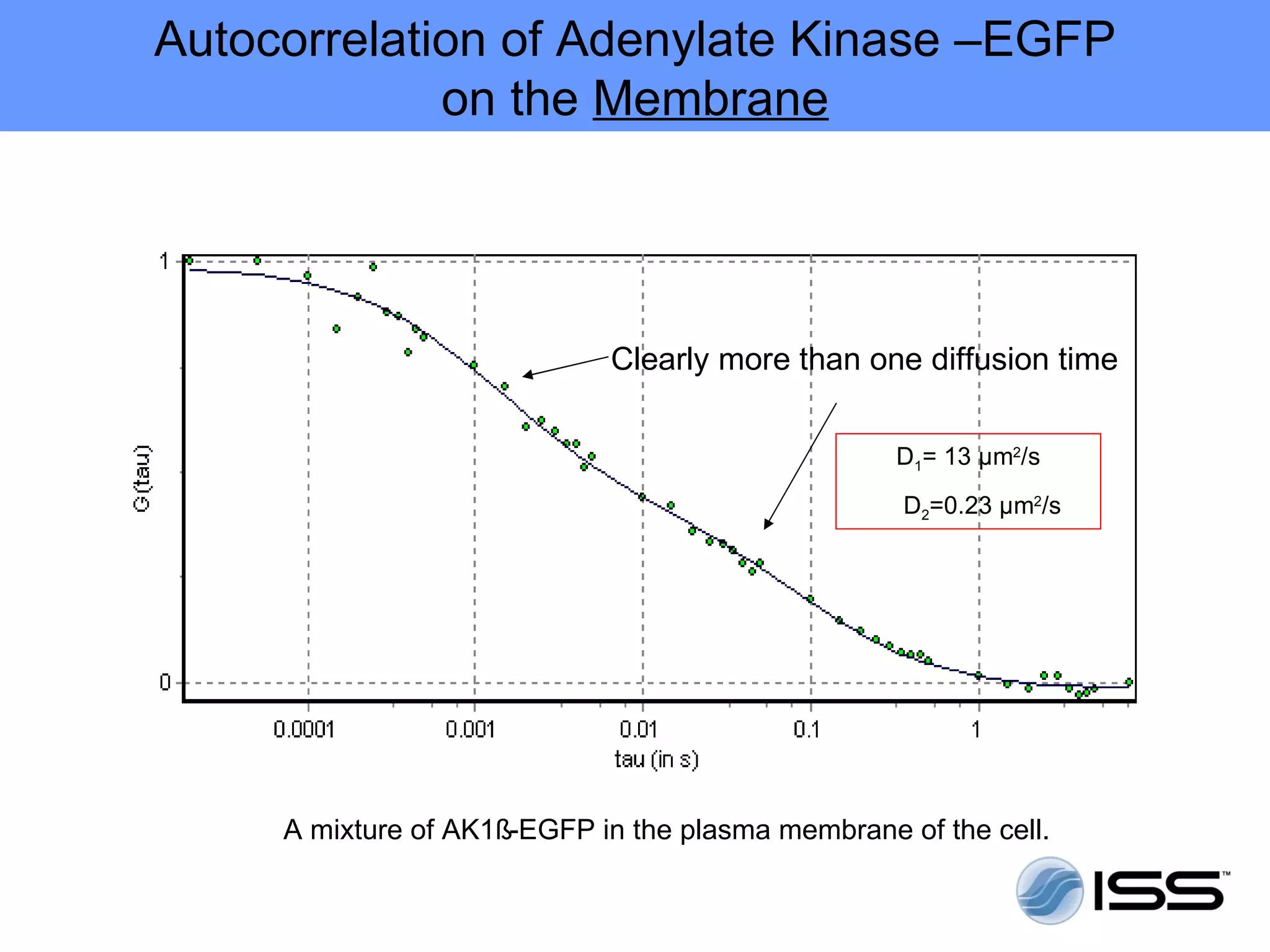
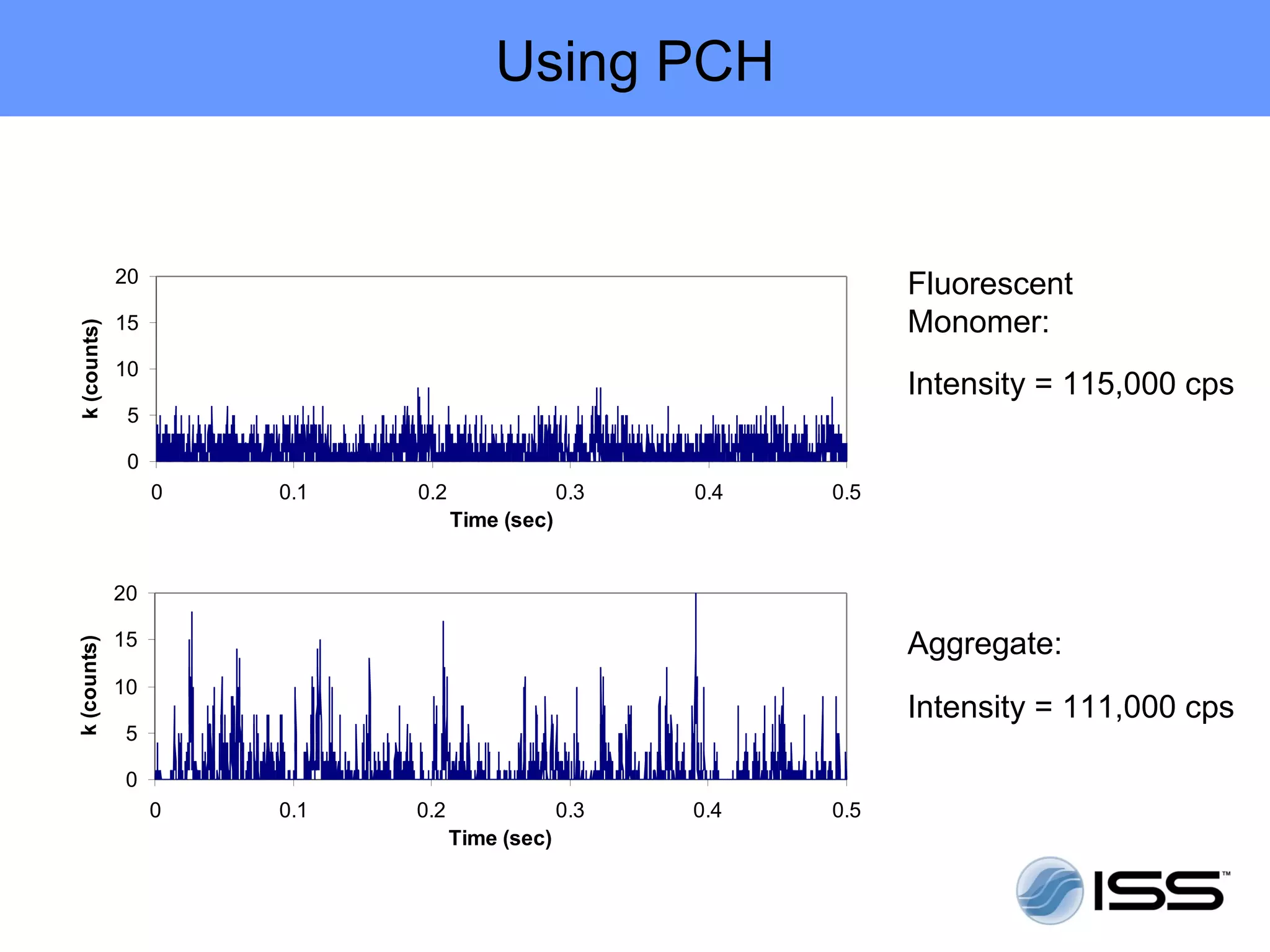
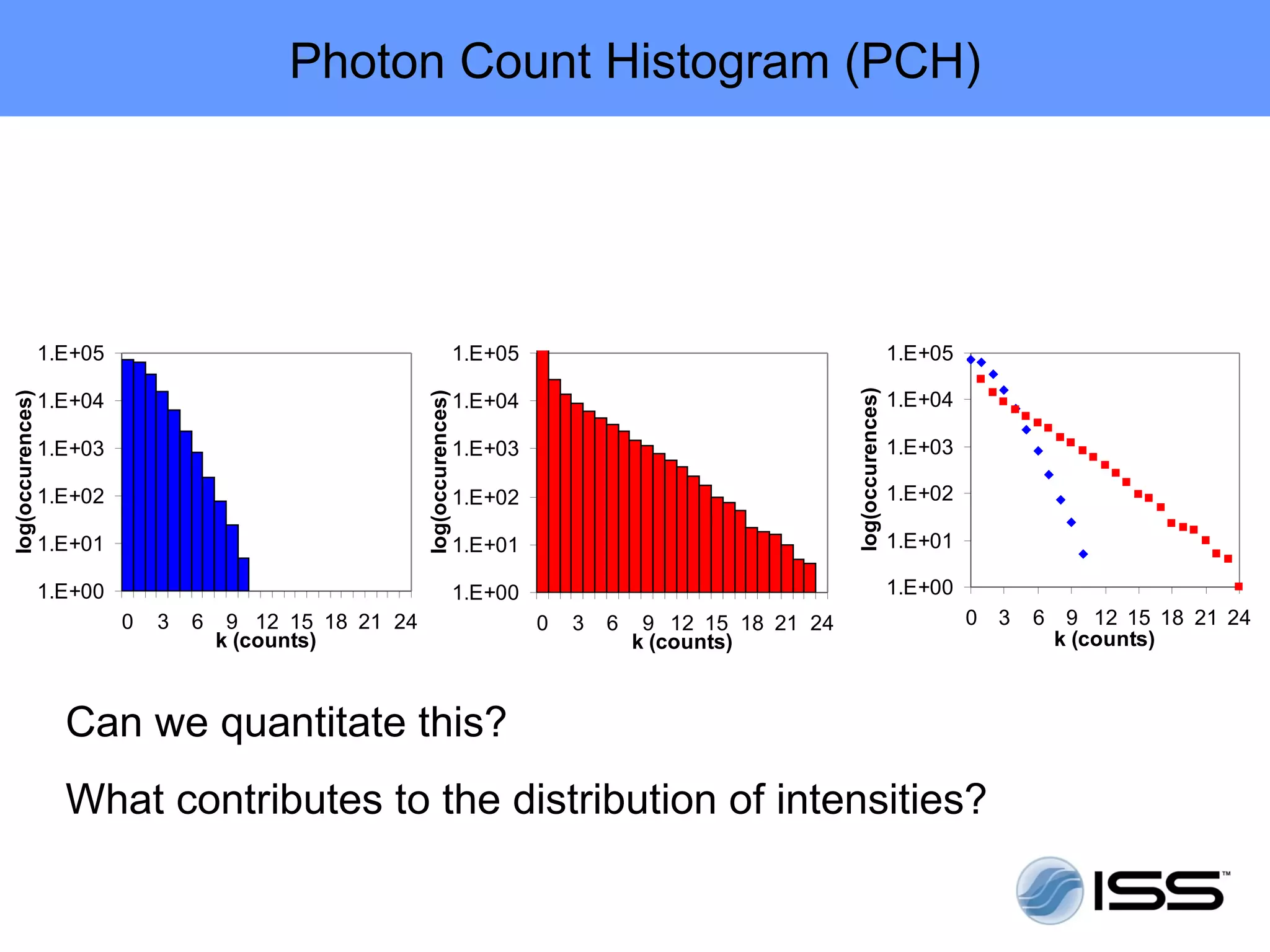
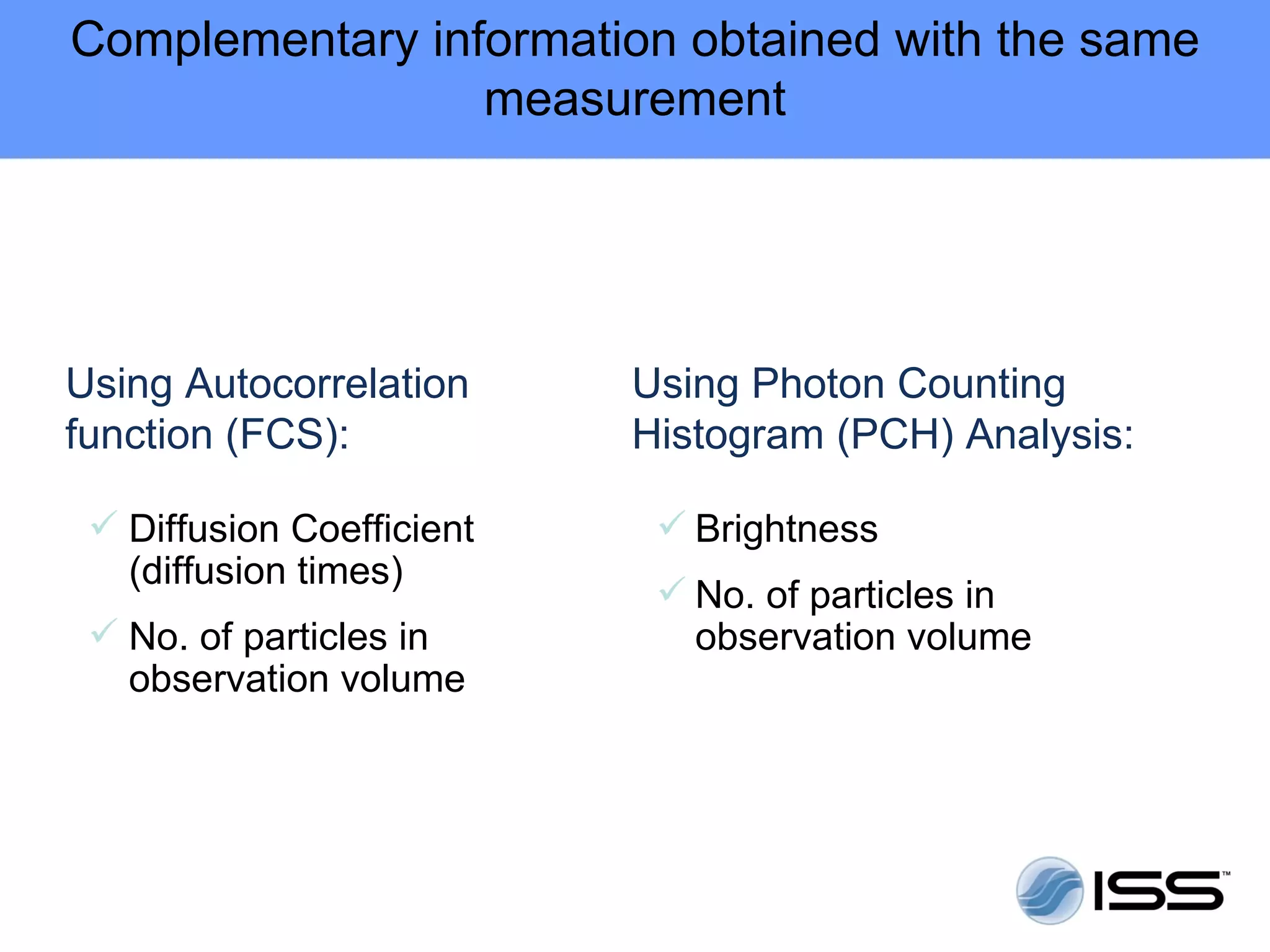
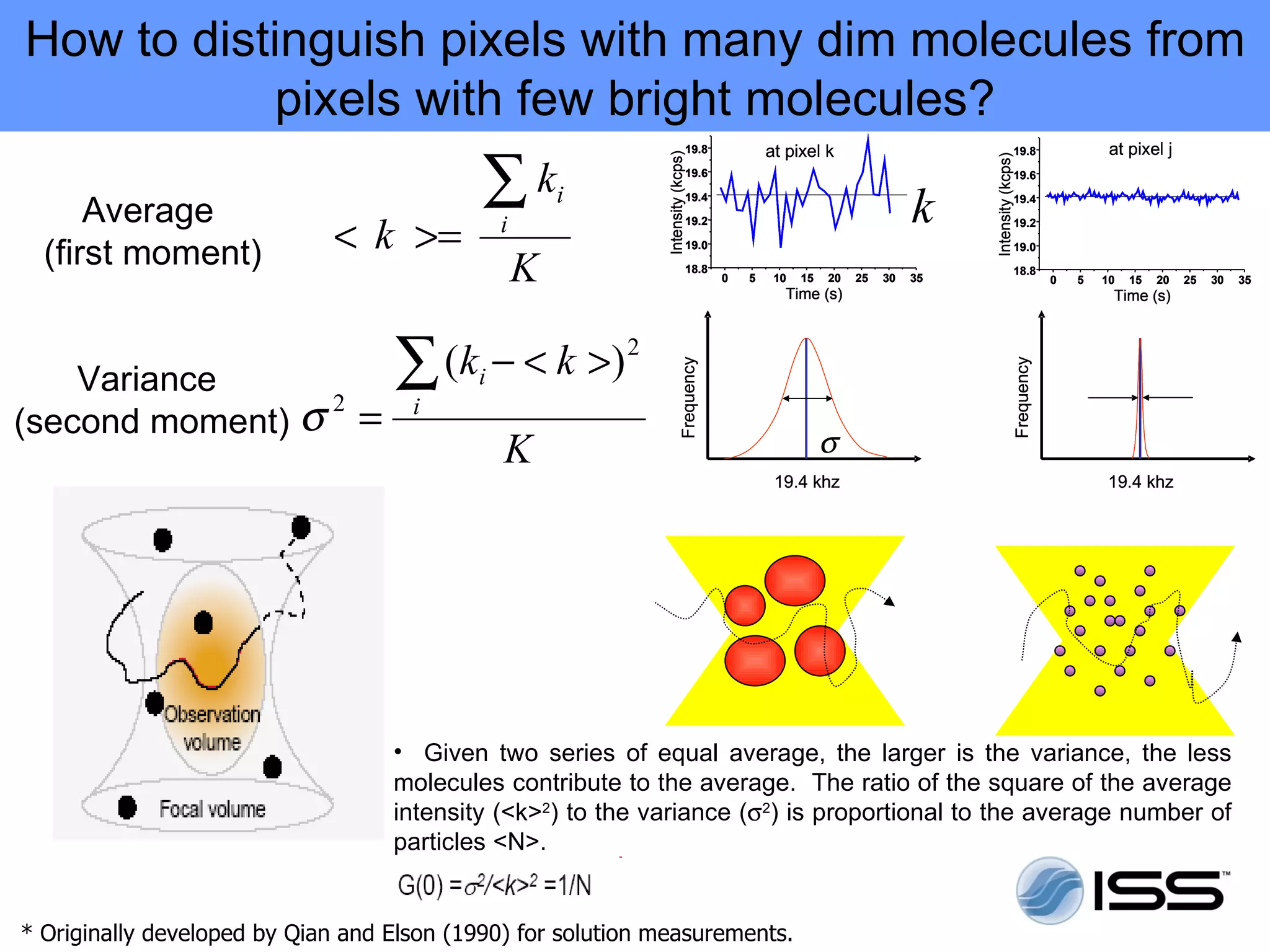
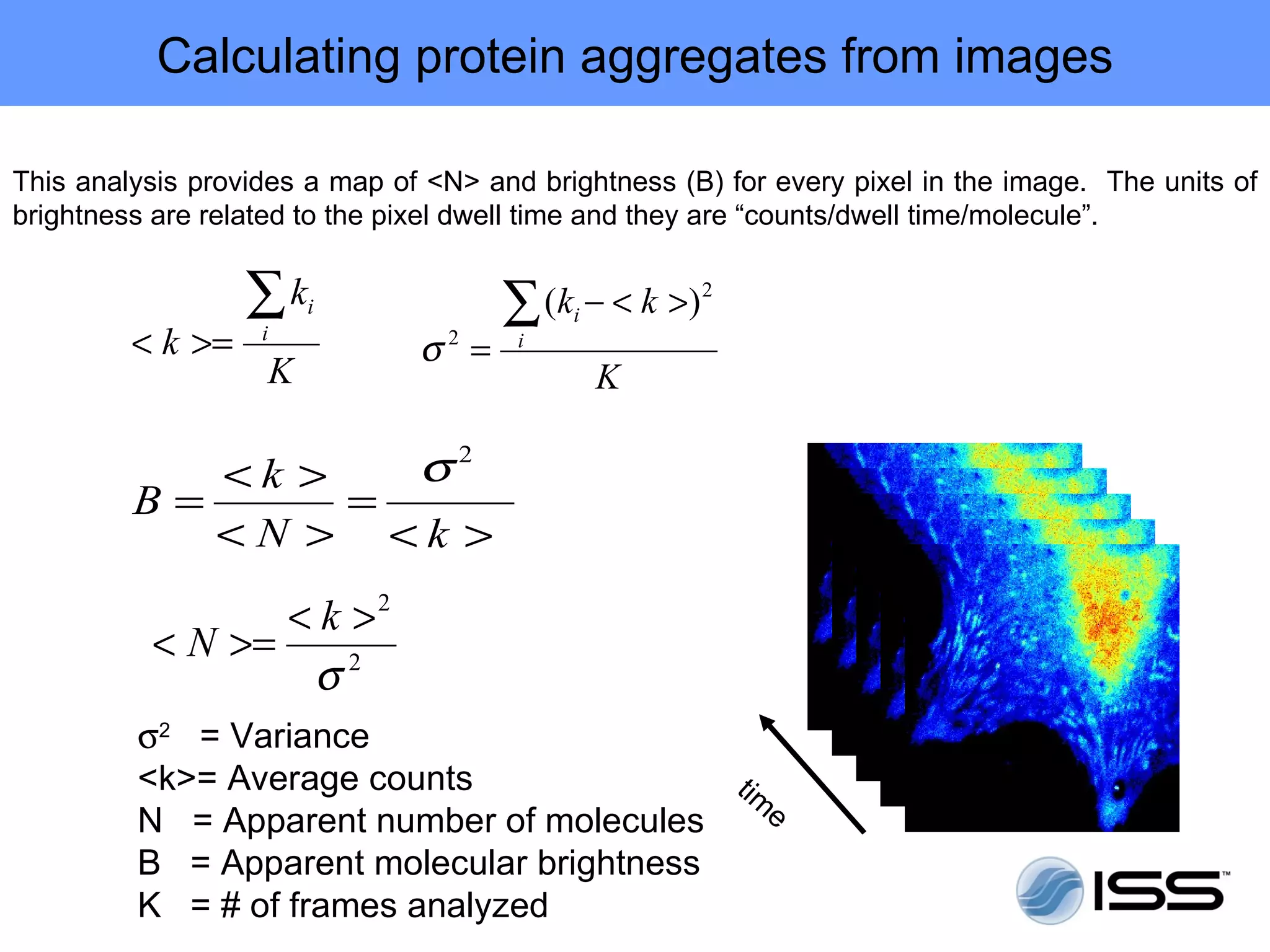
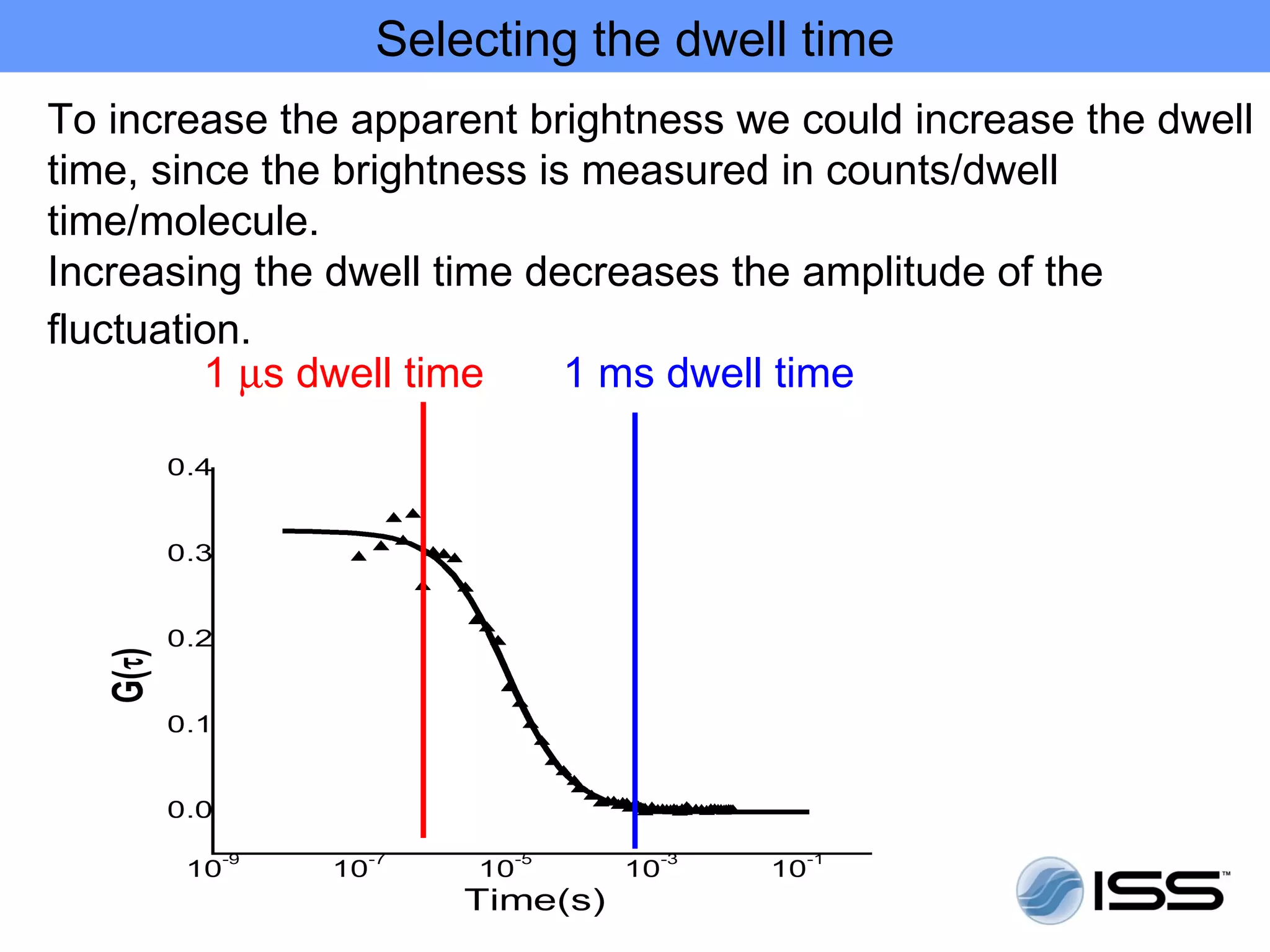
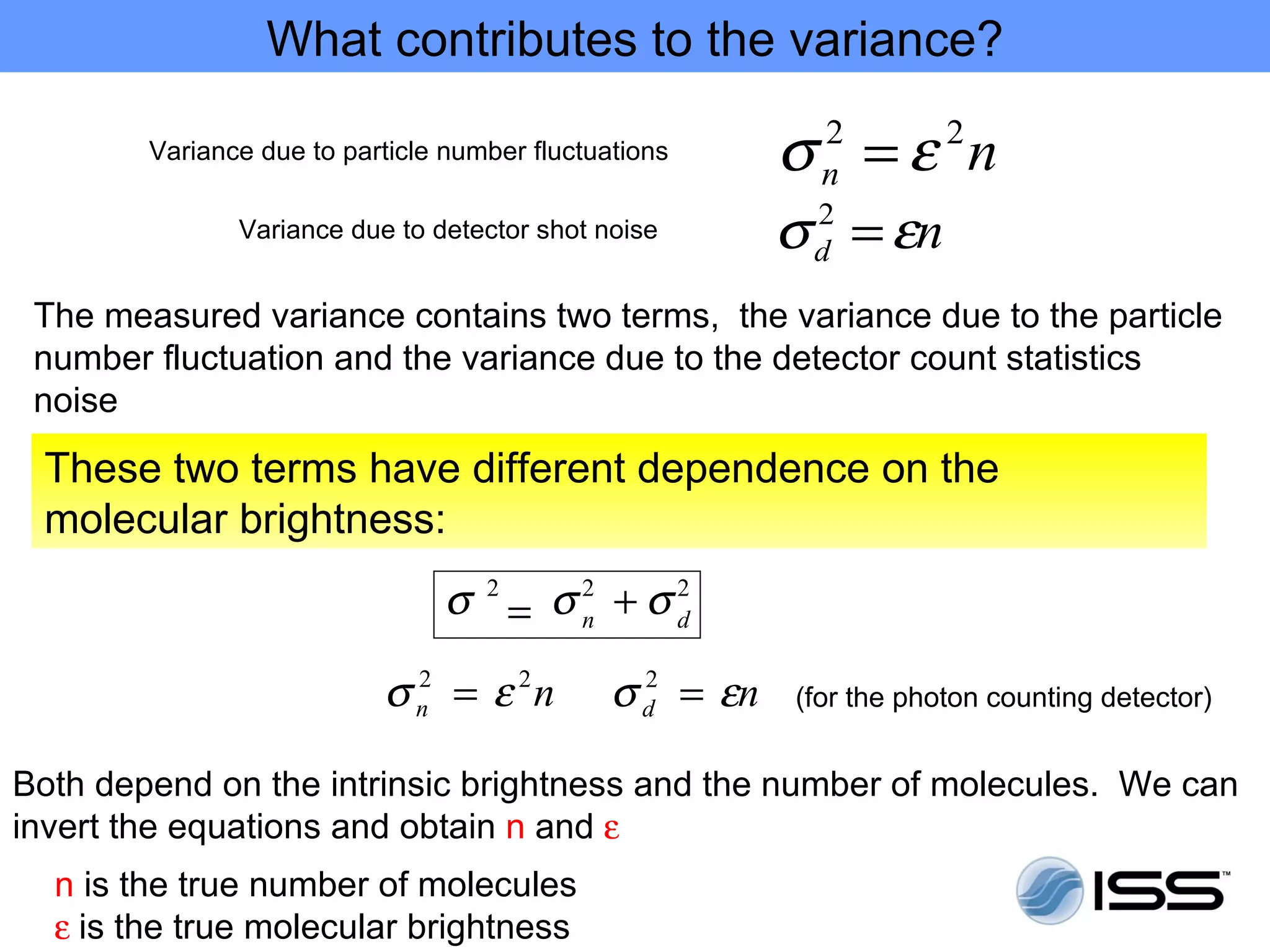
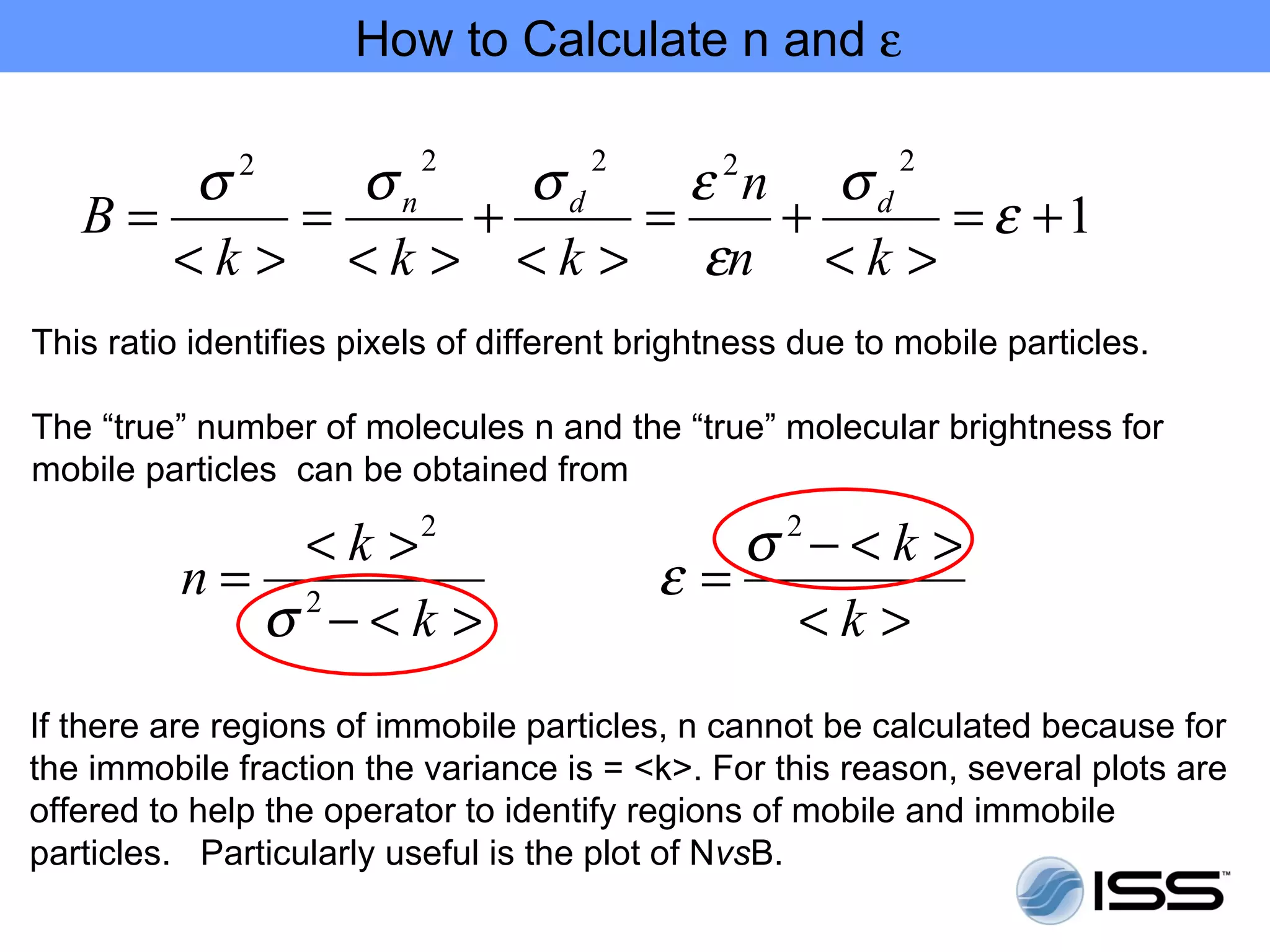
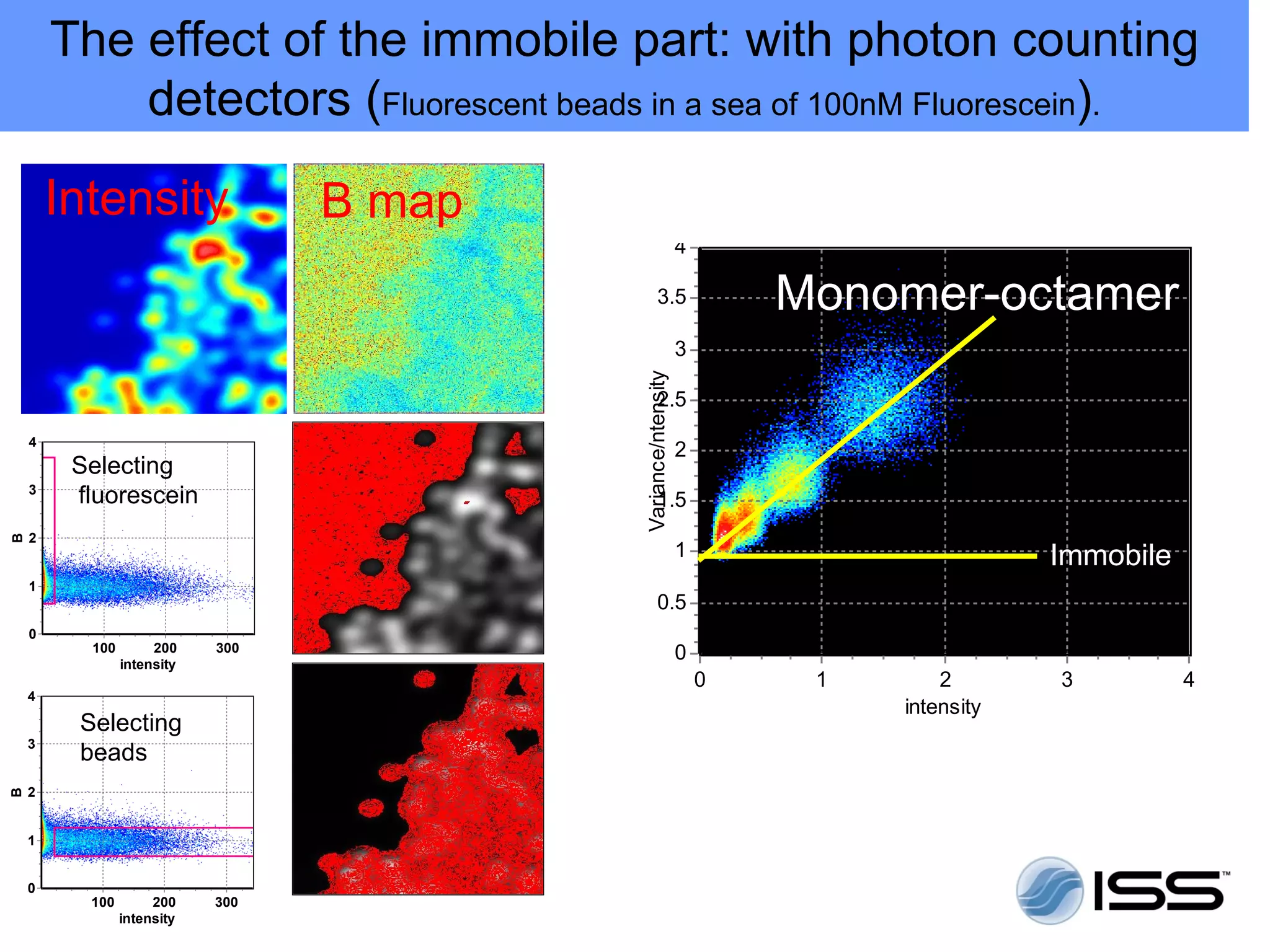
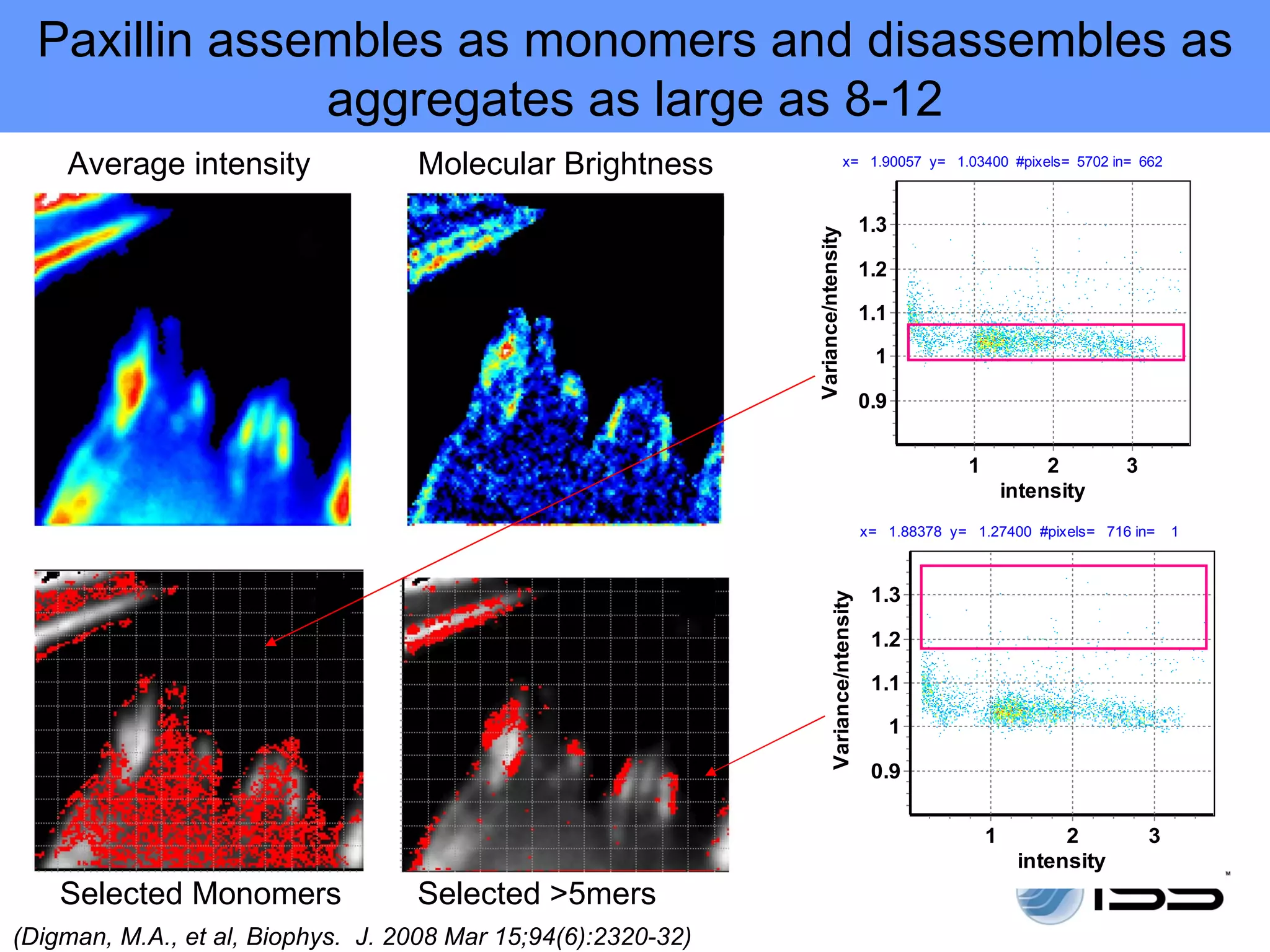
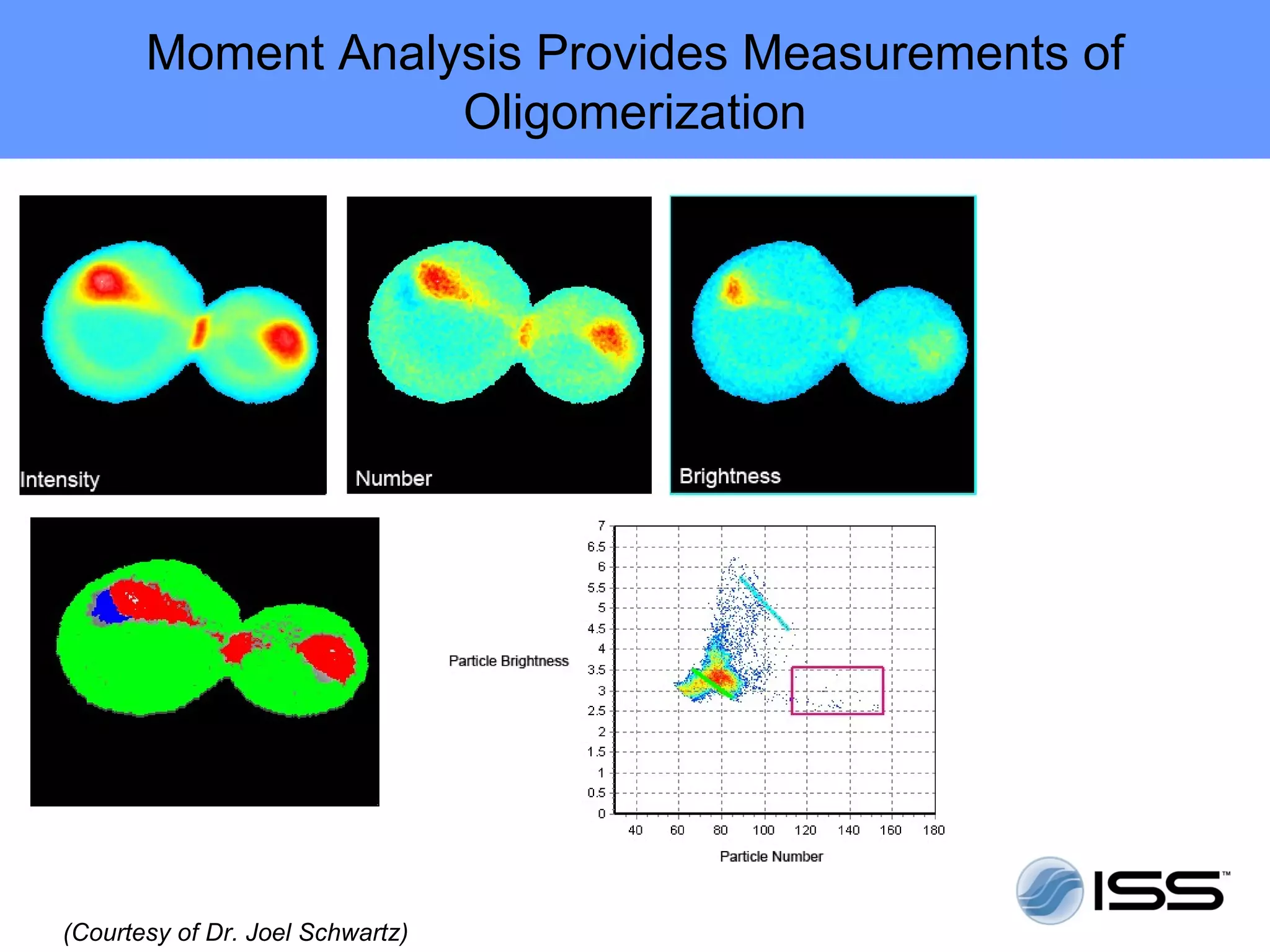
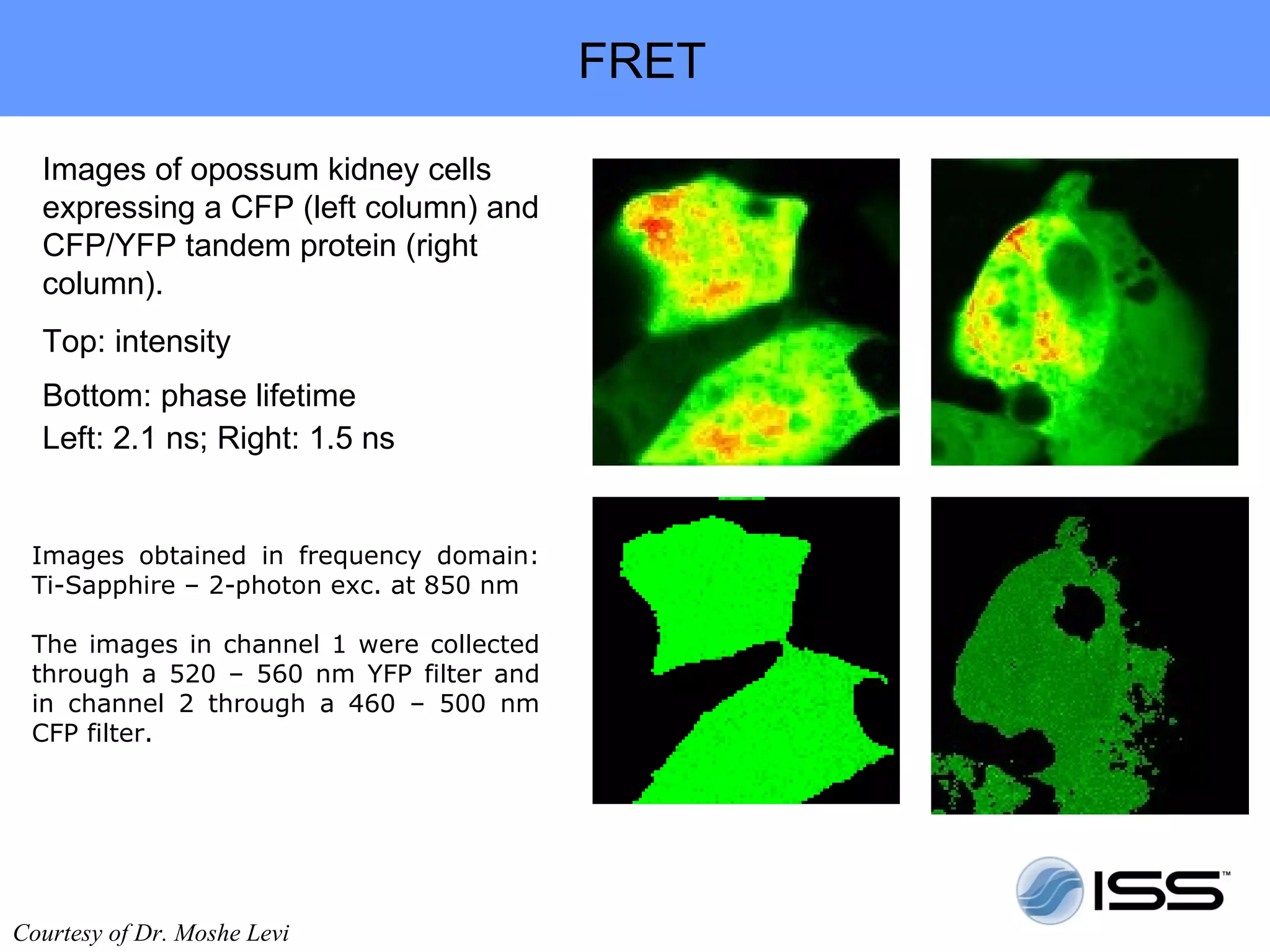
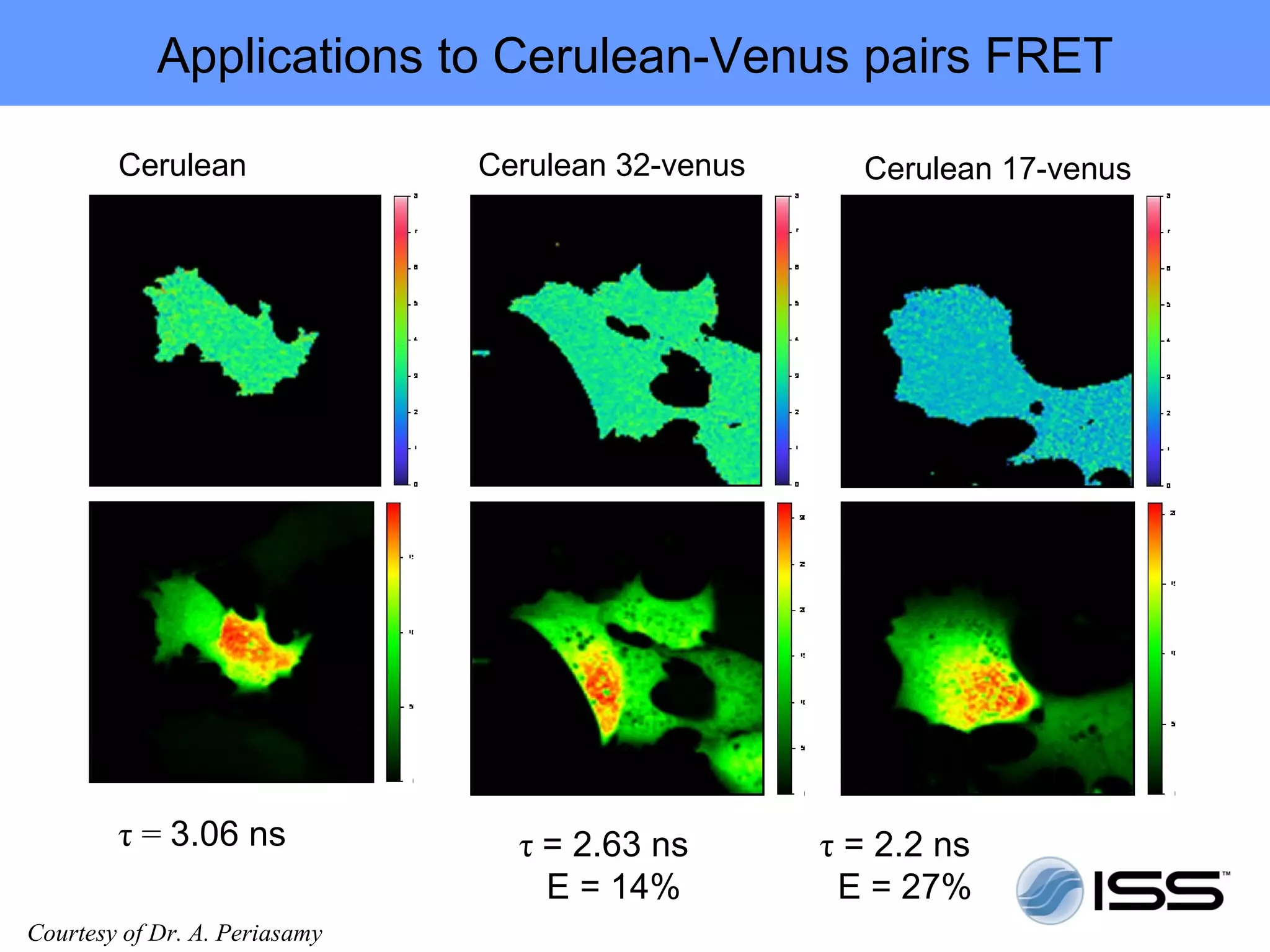
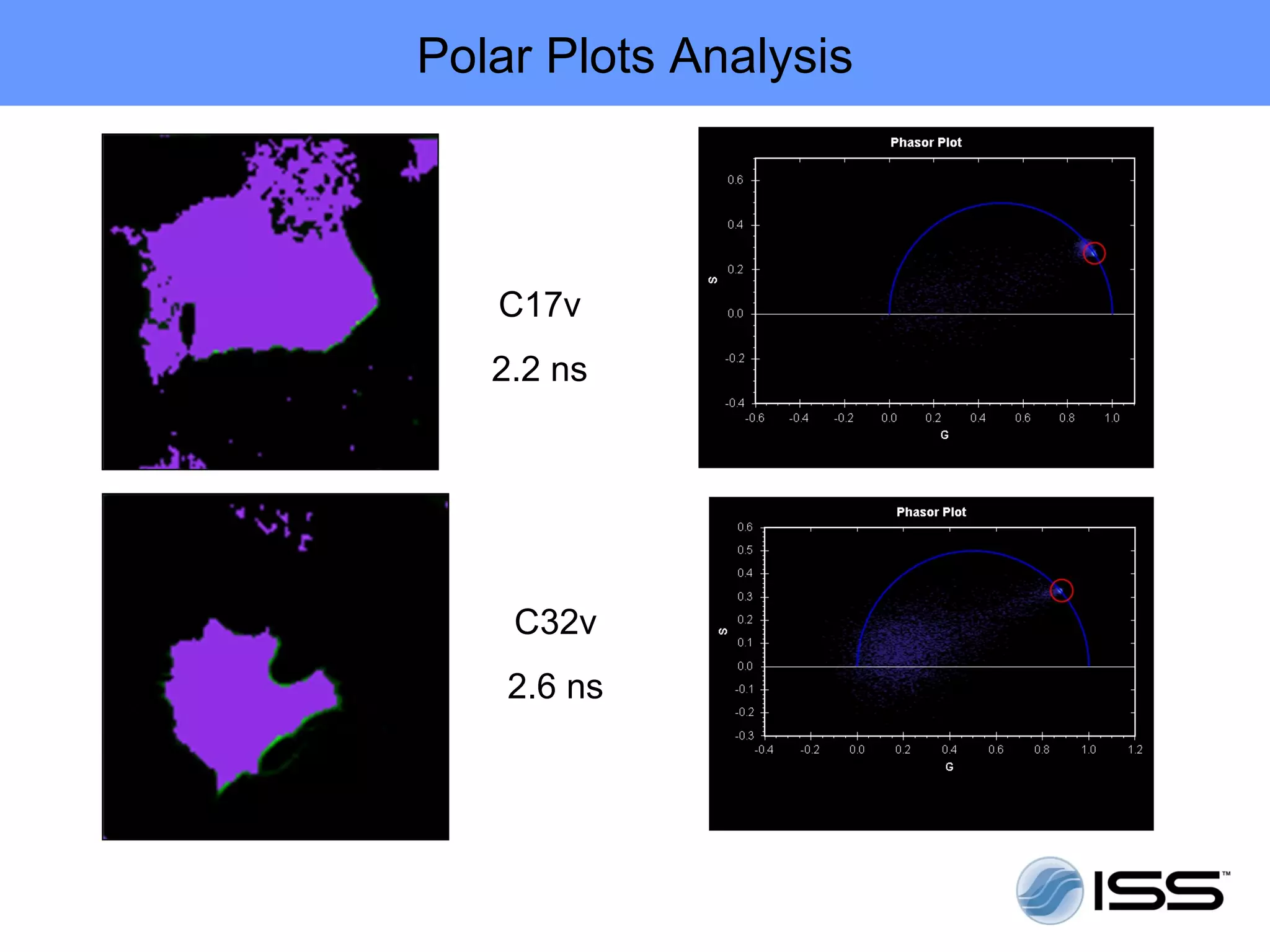
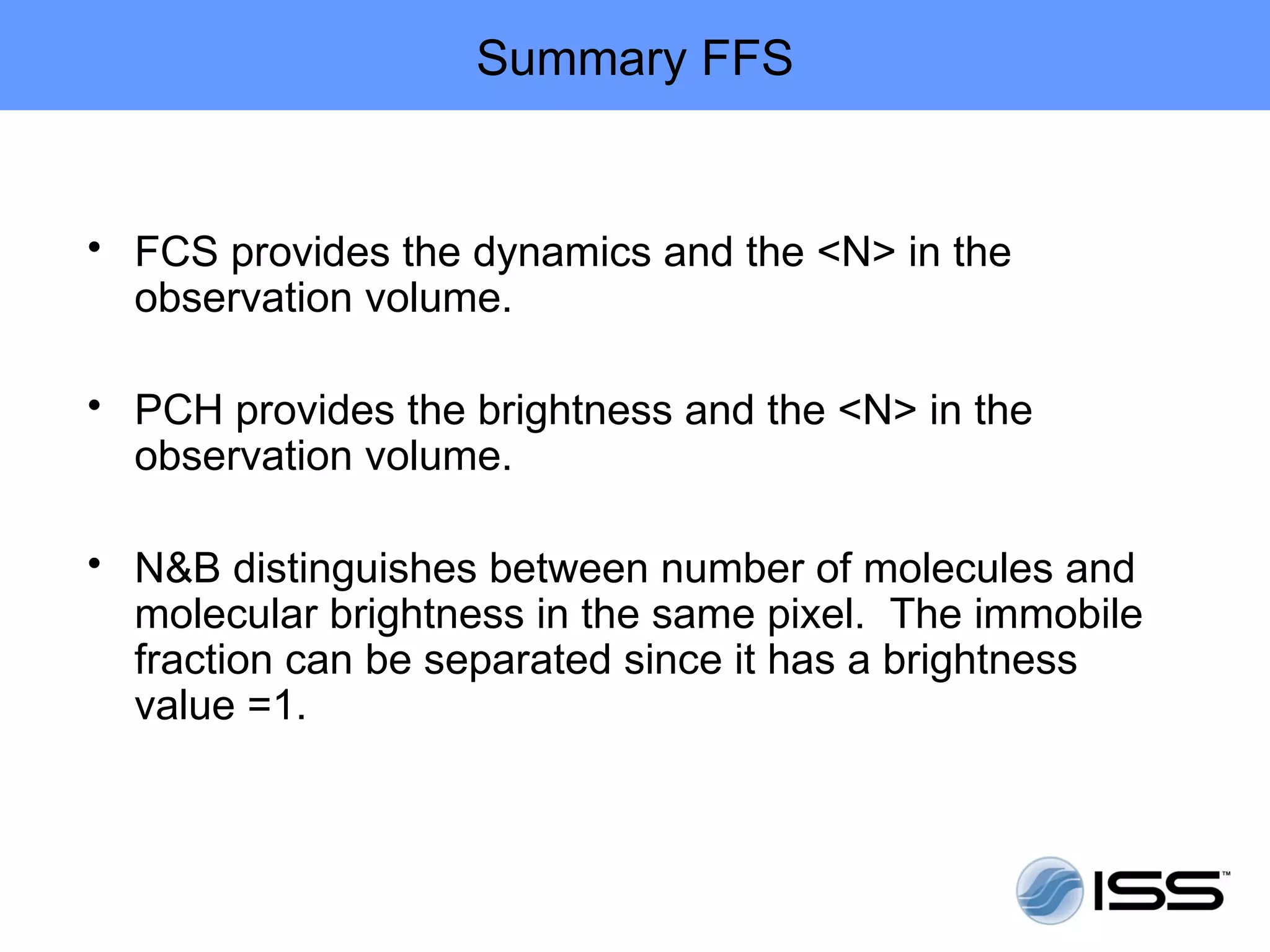
The document discusses fluorescence fluctuations spectroscopy (FFS) techniques such as fluorescence correlation spectroscopy (FCS) and photon counting histogram (PCH) analysis. FFS can provide information about molecular diffusion, aggregation, and concentration within cells by analyzing fluctuations in fluorescence intensity over time at the microscopic level. Specifically, FFS techniques measure fluorescence intensity autocorrelation and distribution to determine numbers of molecules, brightness, and diffusion coefficients. Related techniques like number and brightness (N&B) analysis provide pixel-level maps of these parameters within cell images. FFS is a powerful tool for studying molecular dynamics and clustering in live cells.