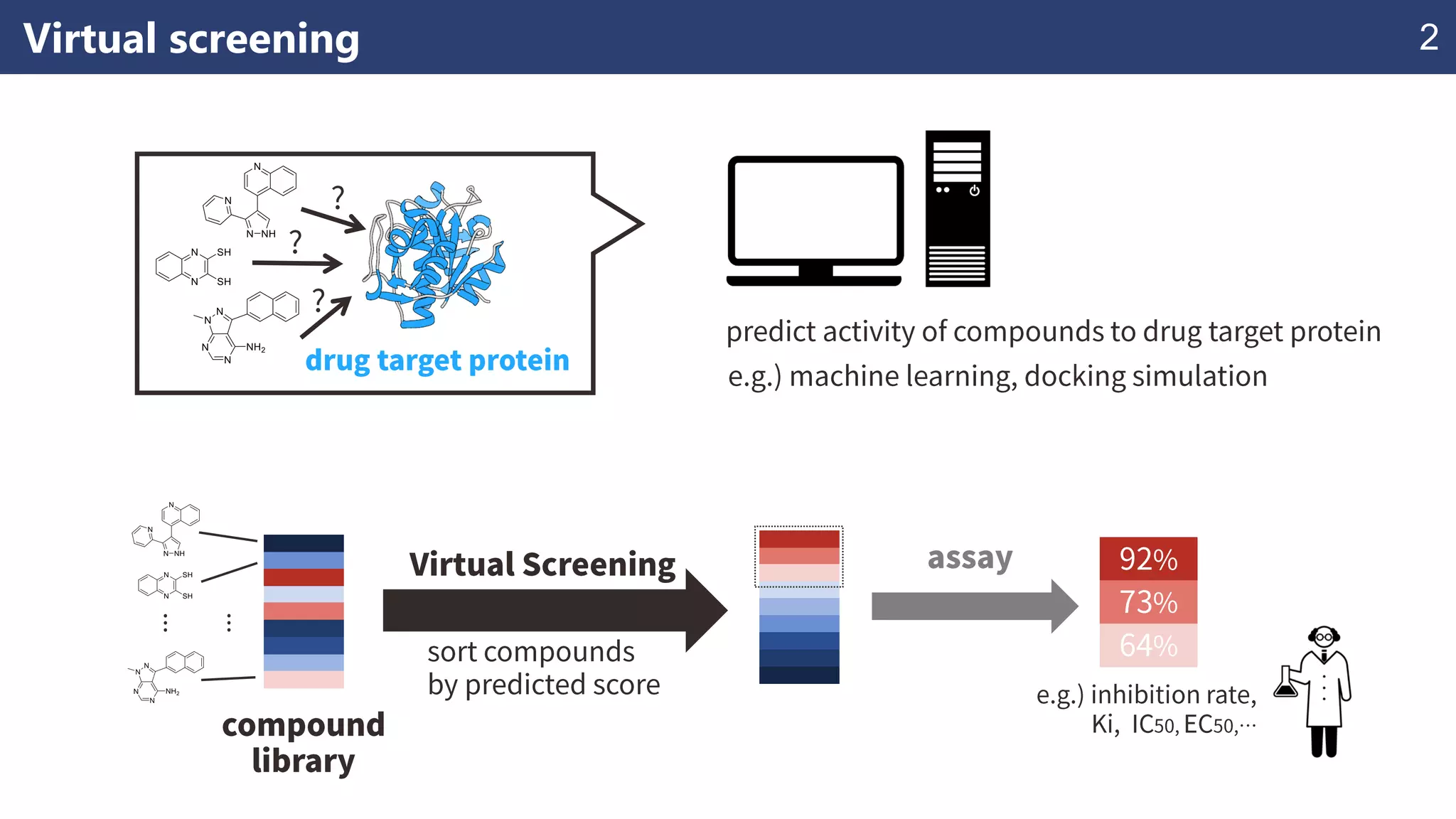
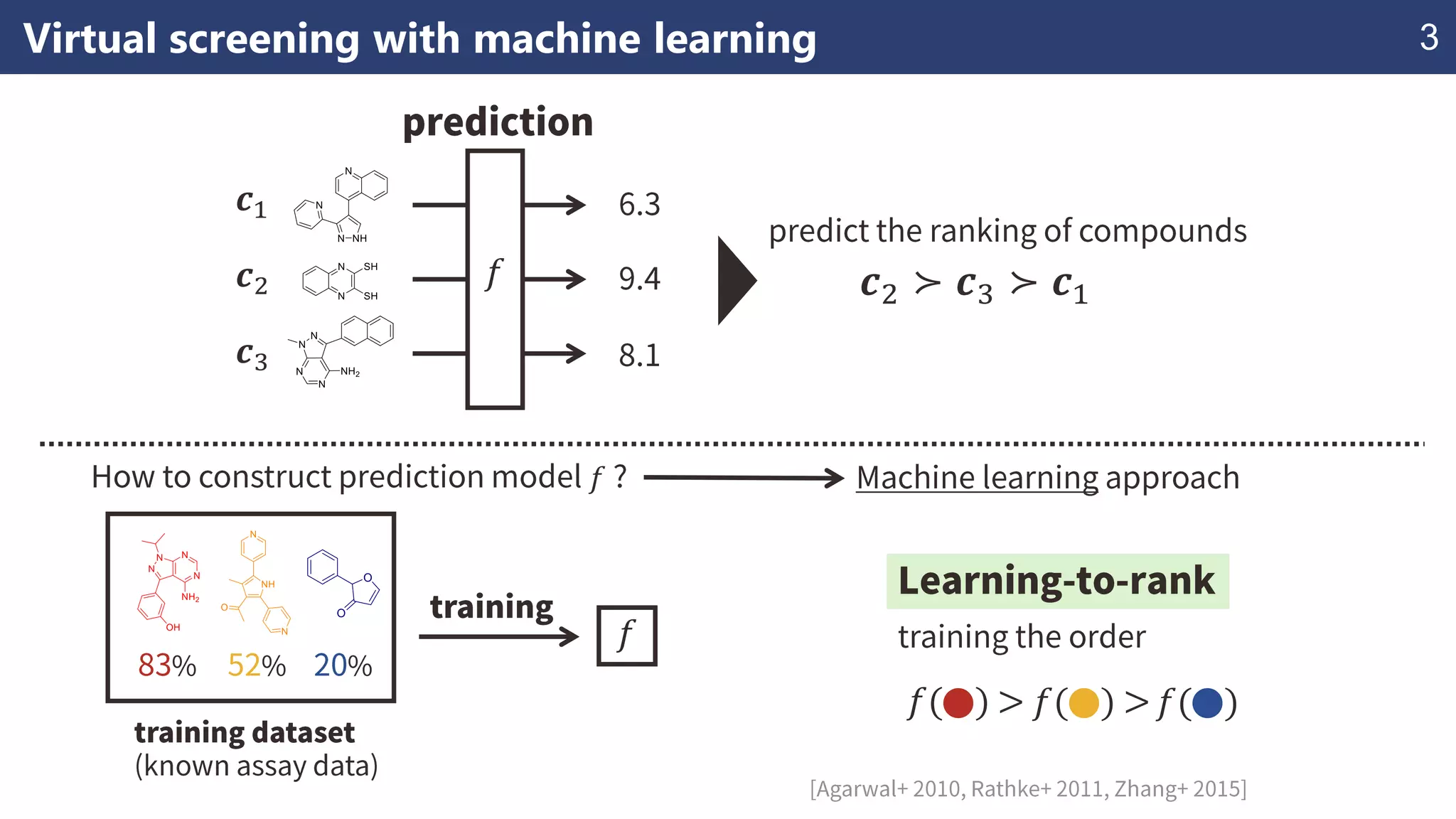
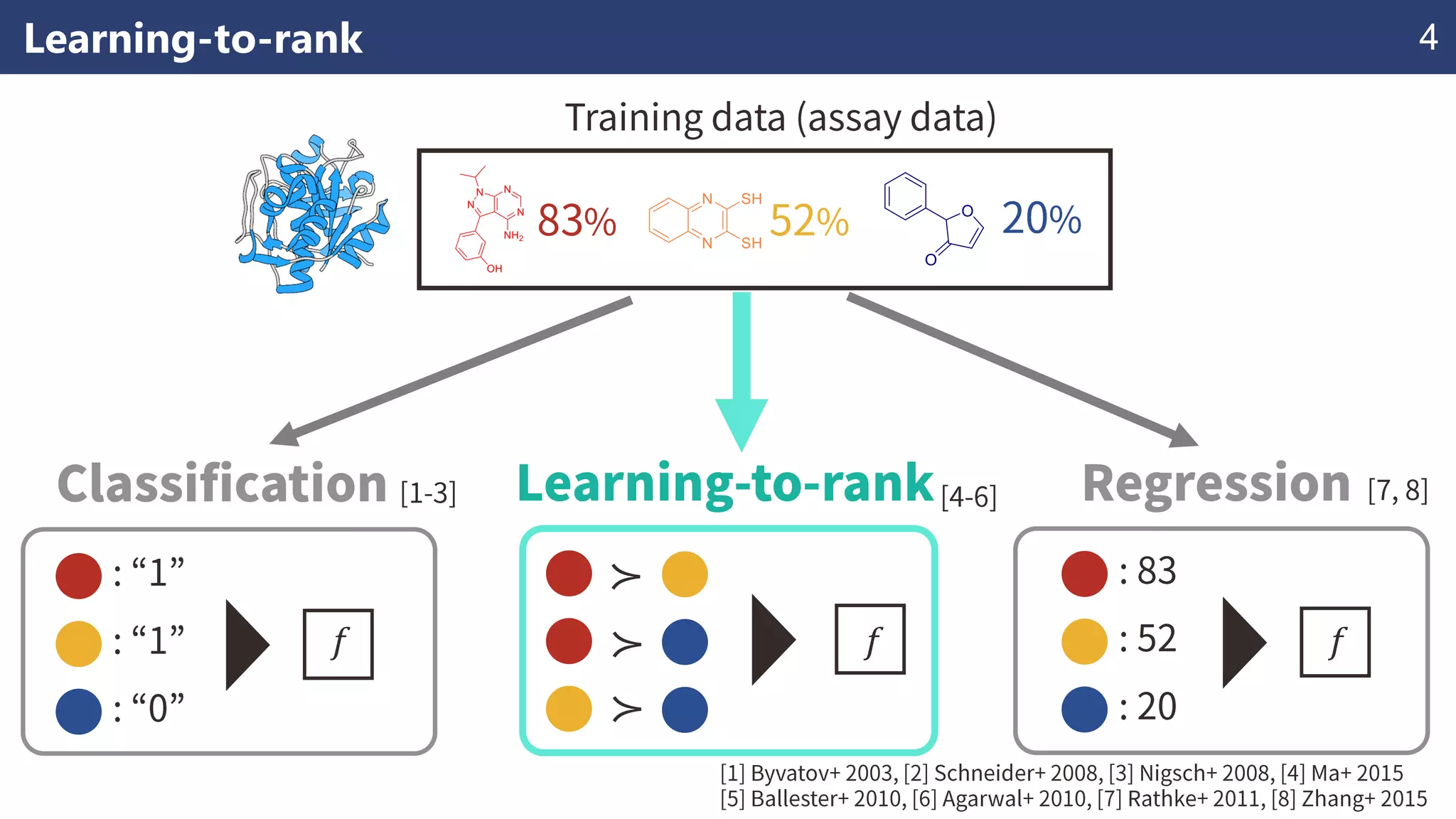
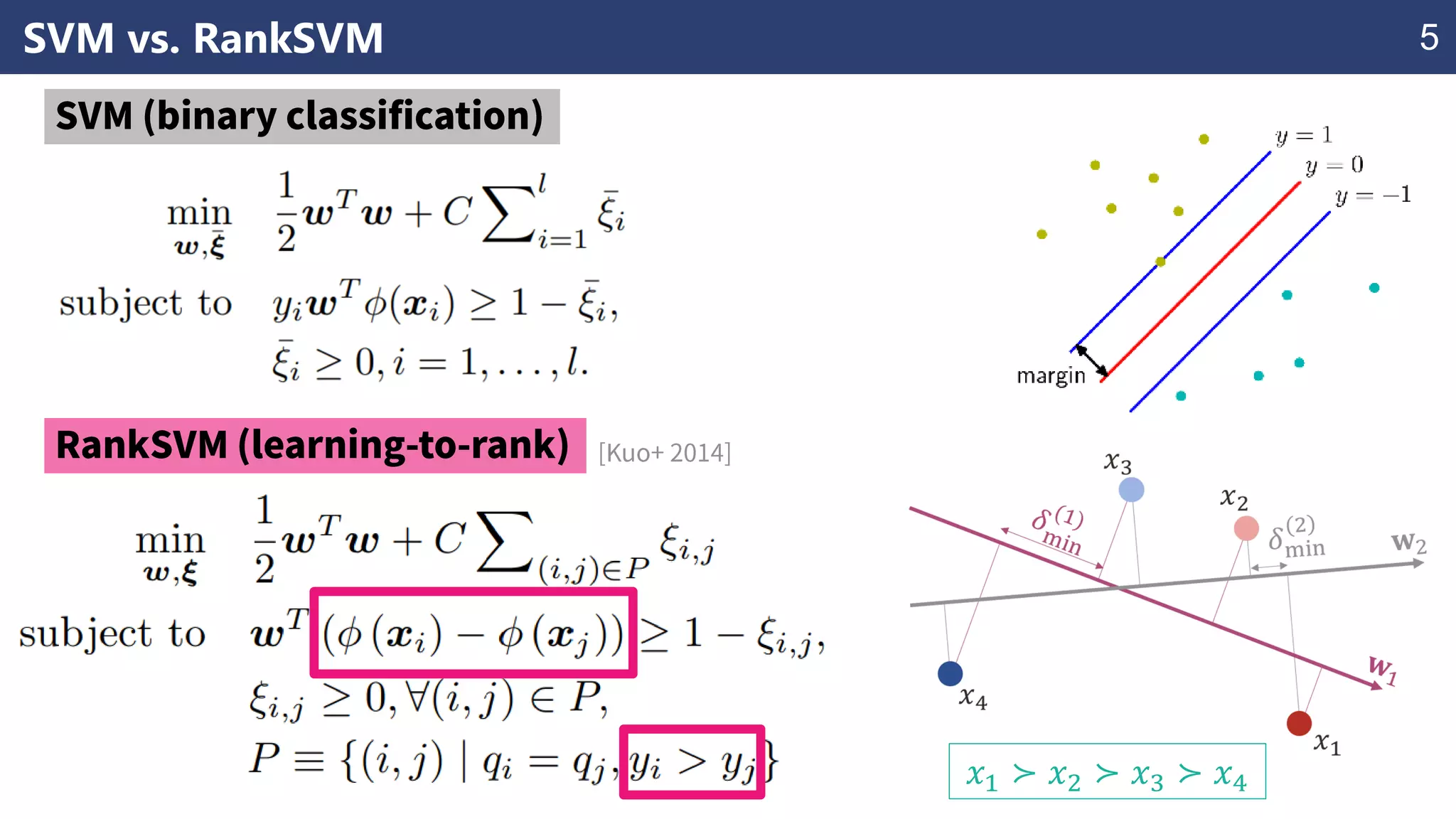
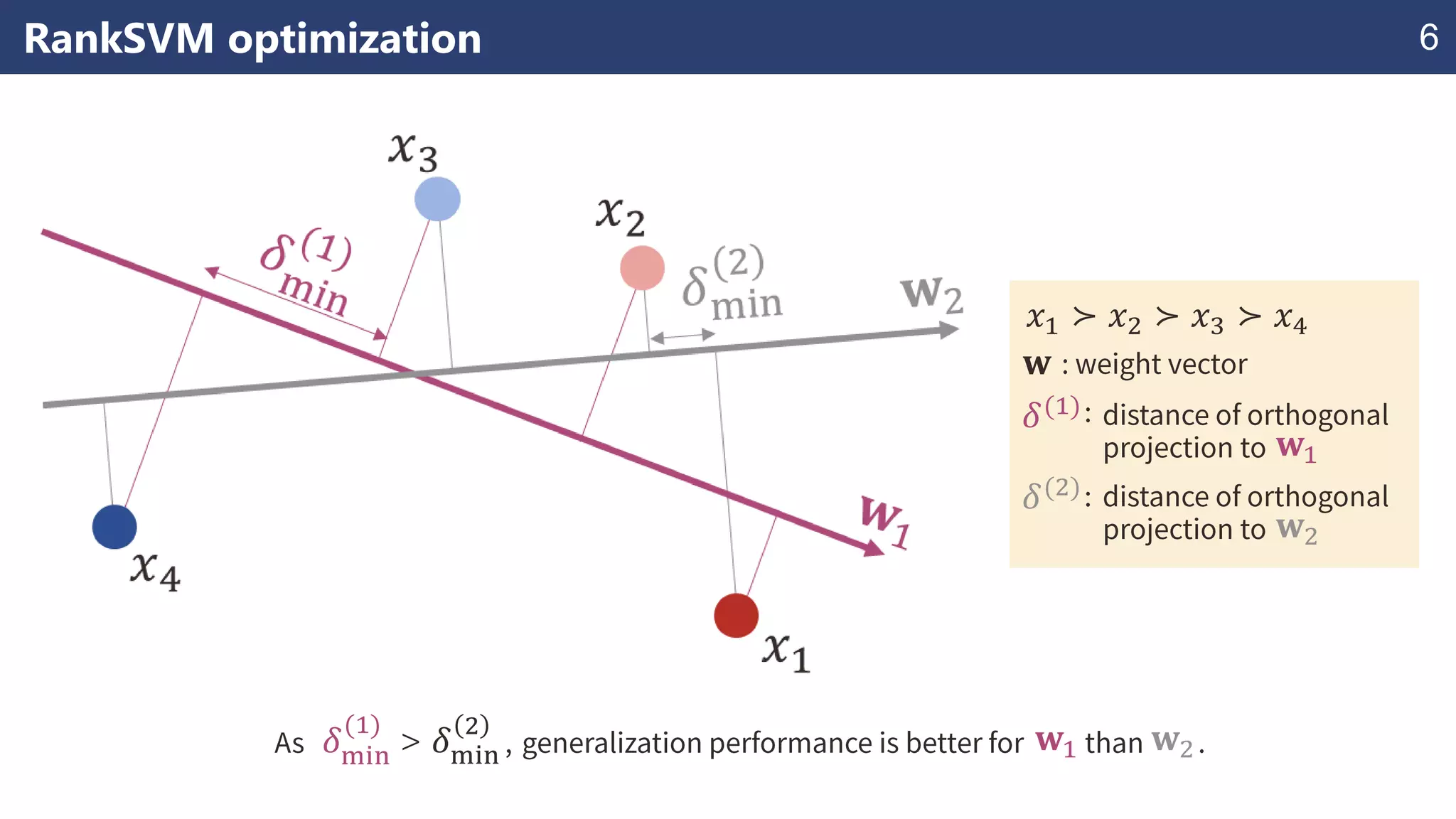
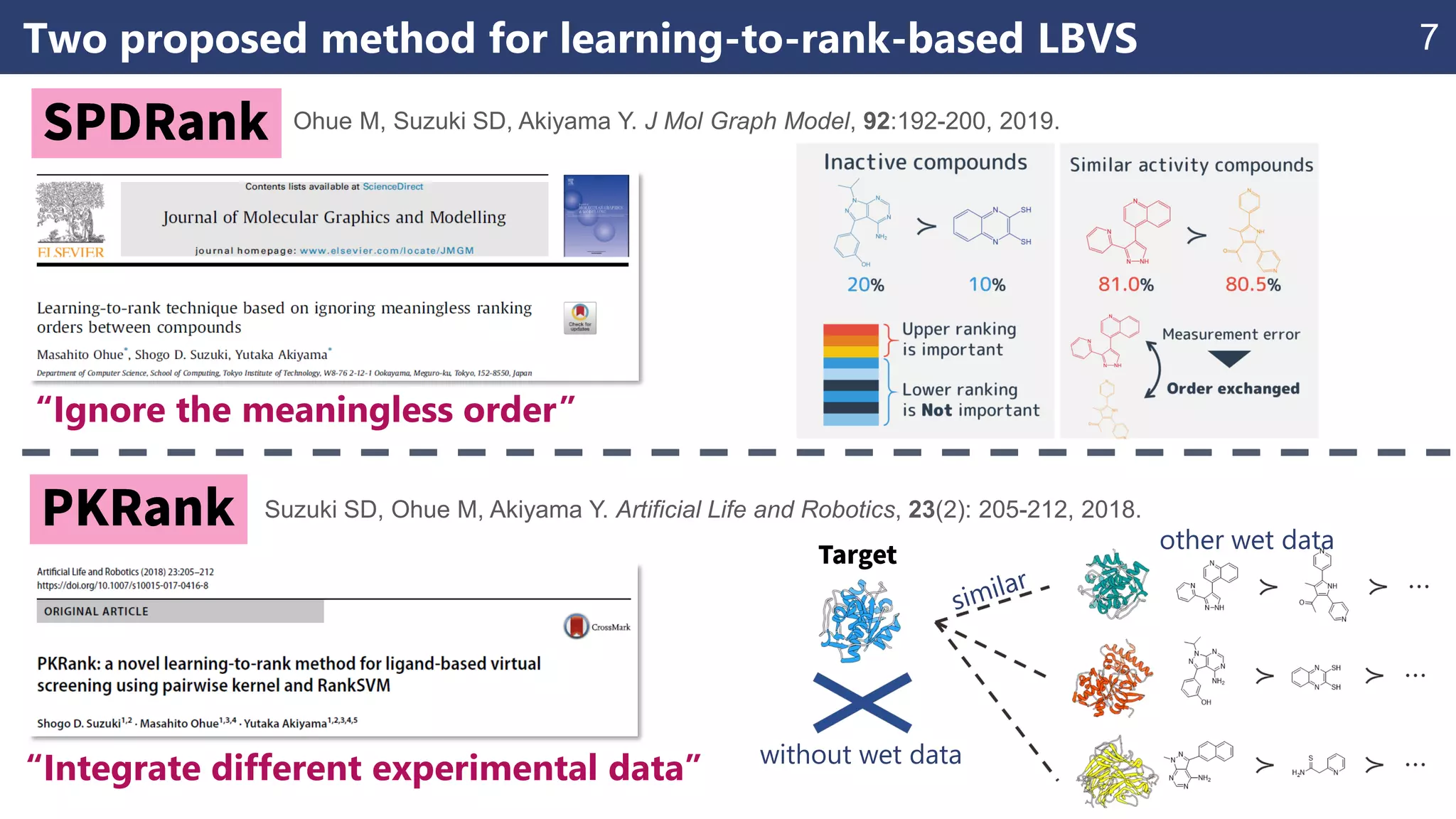
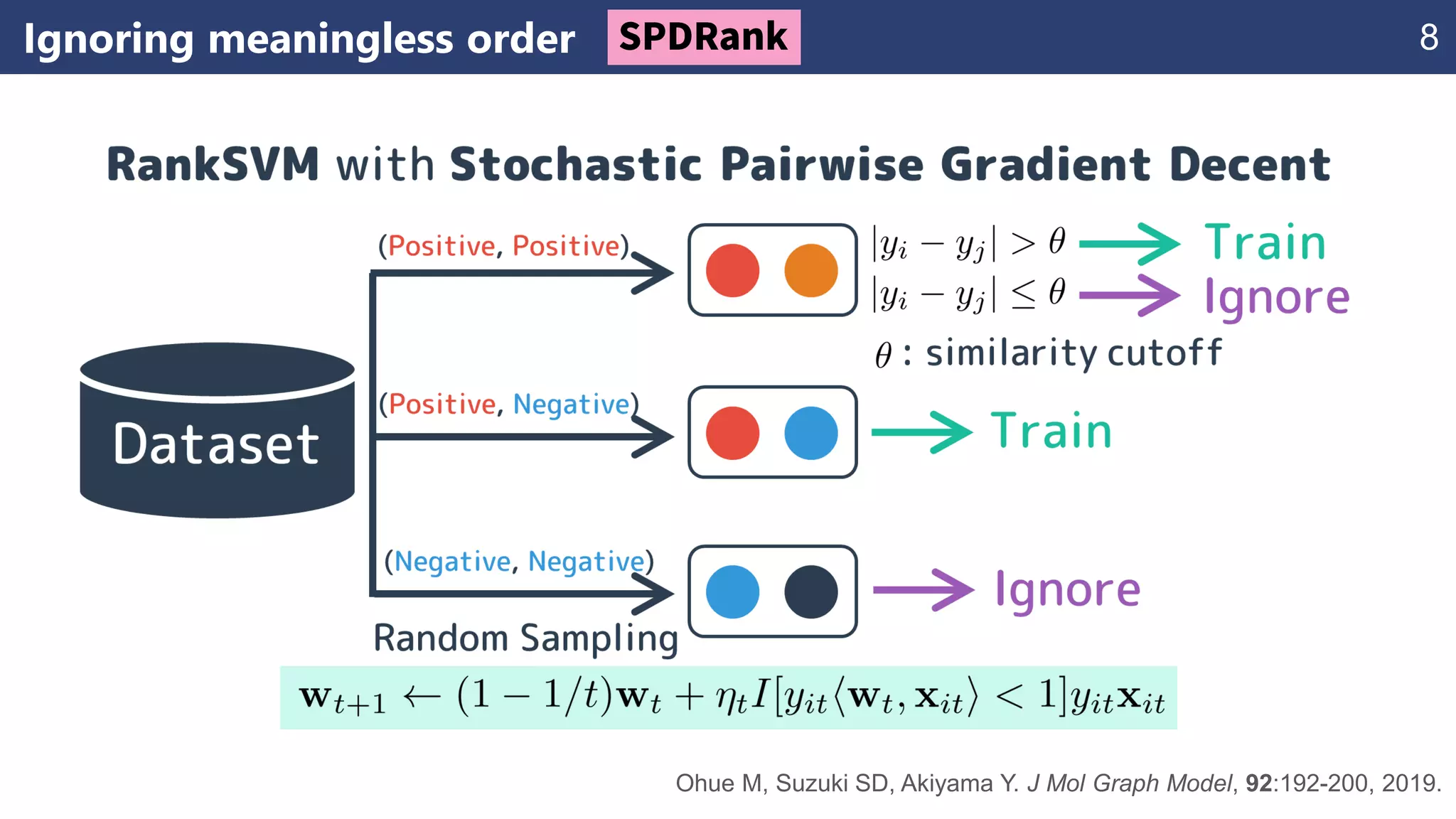
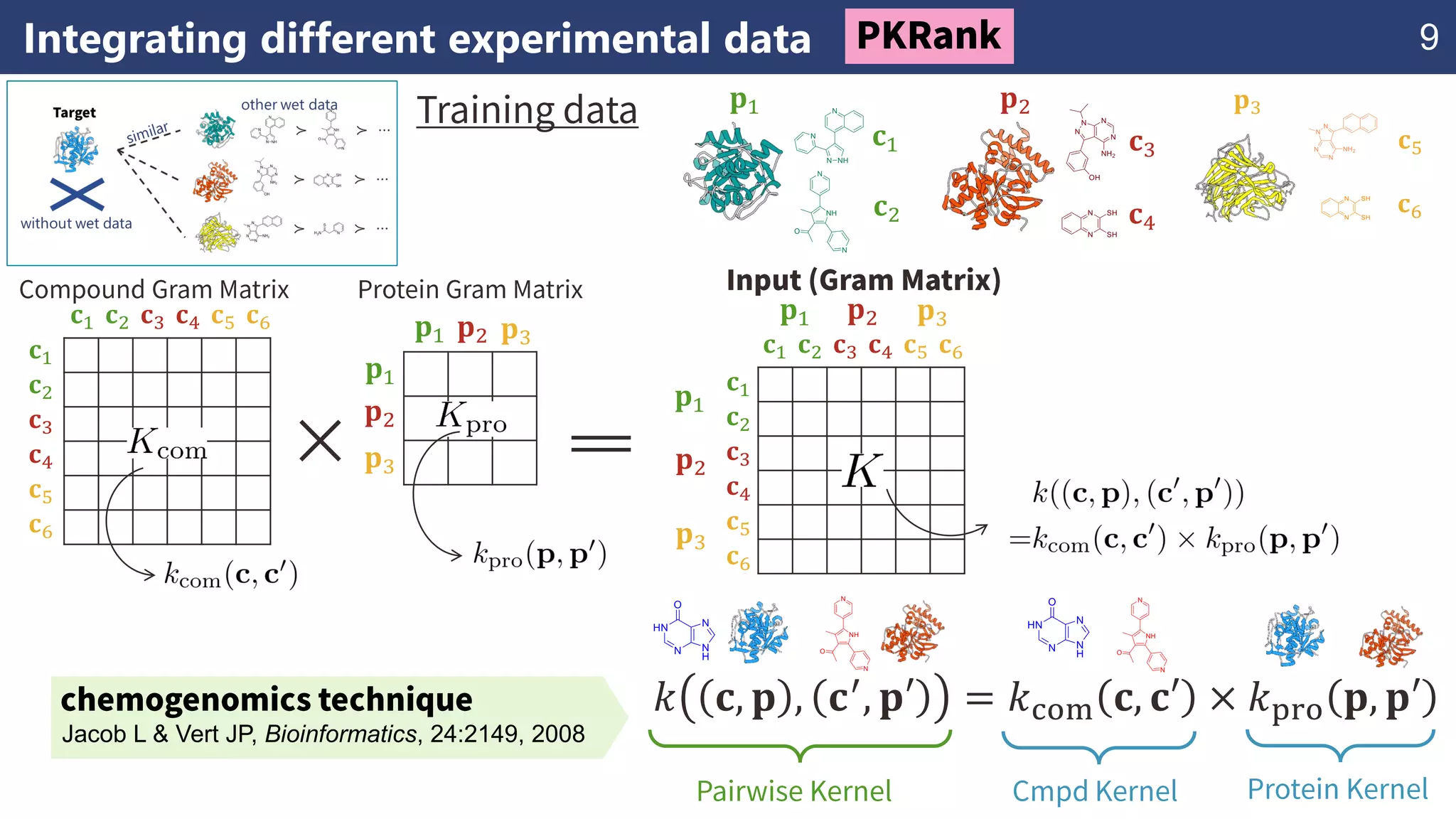
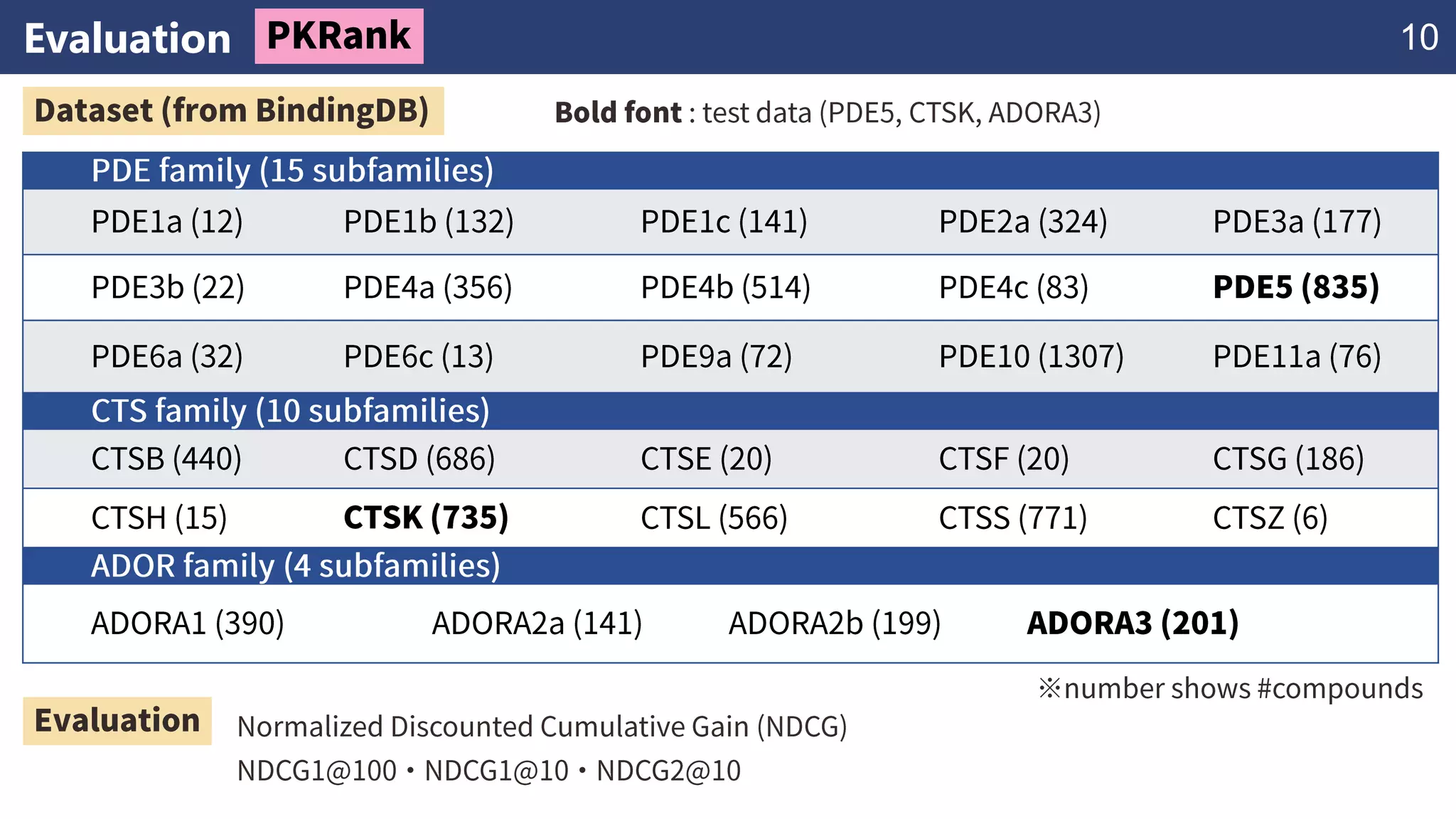
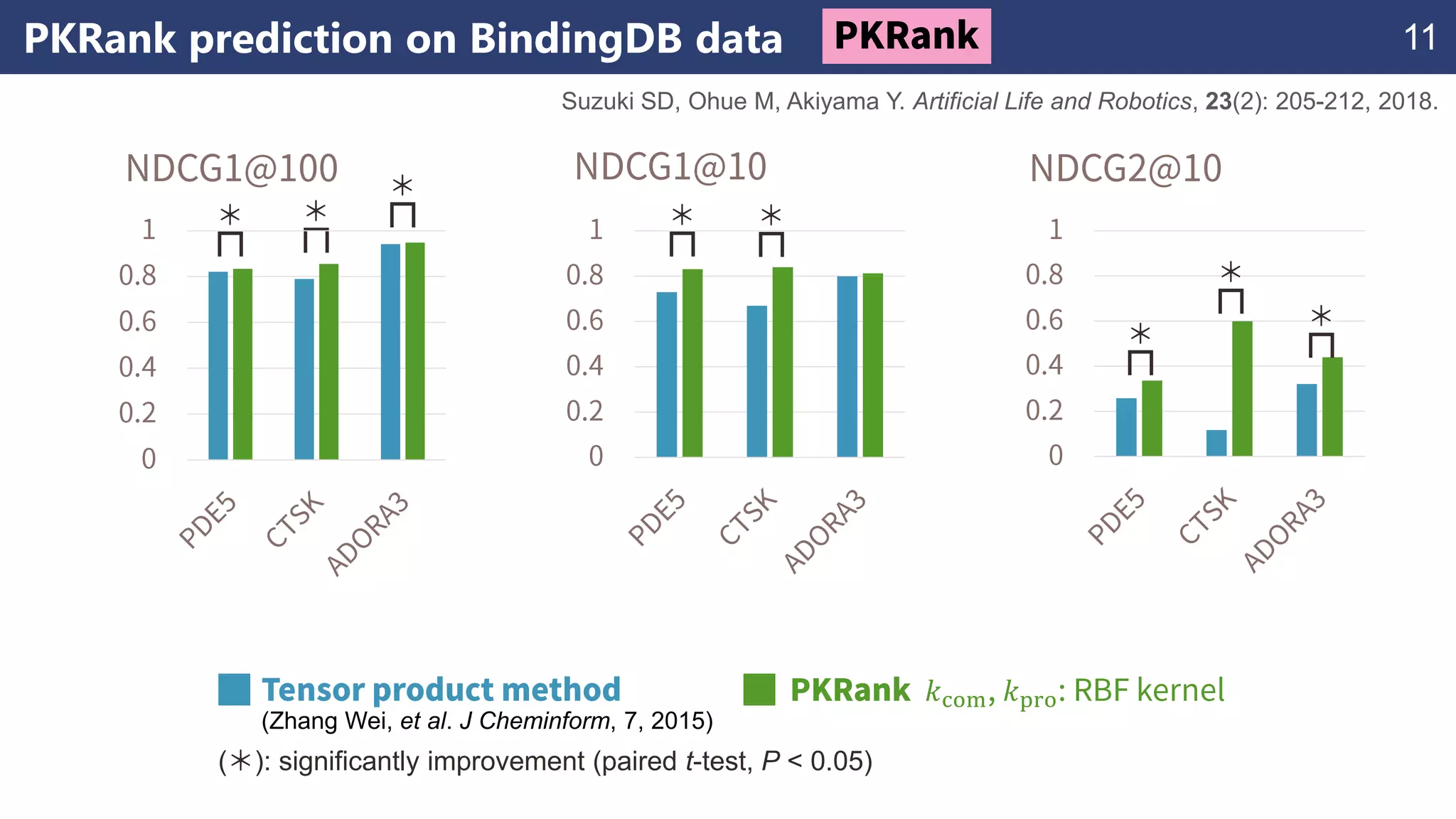
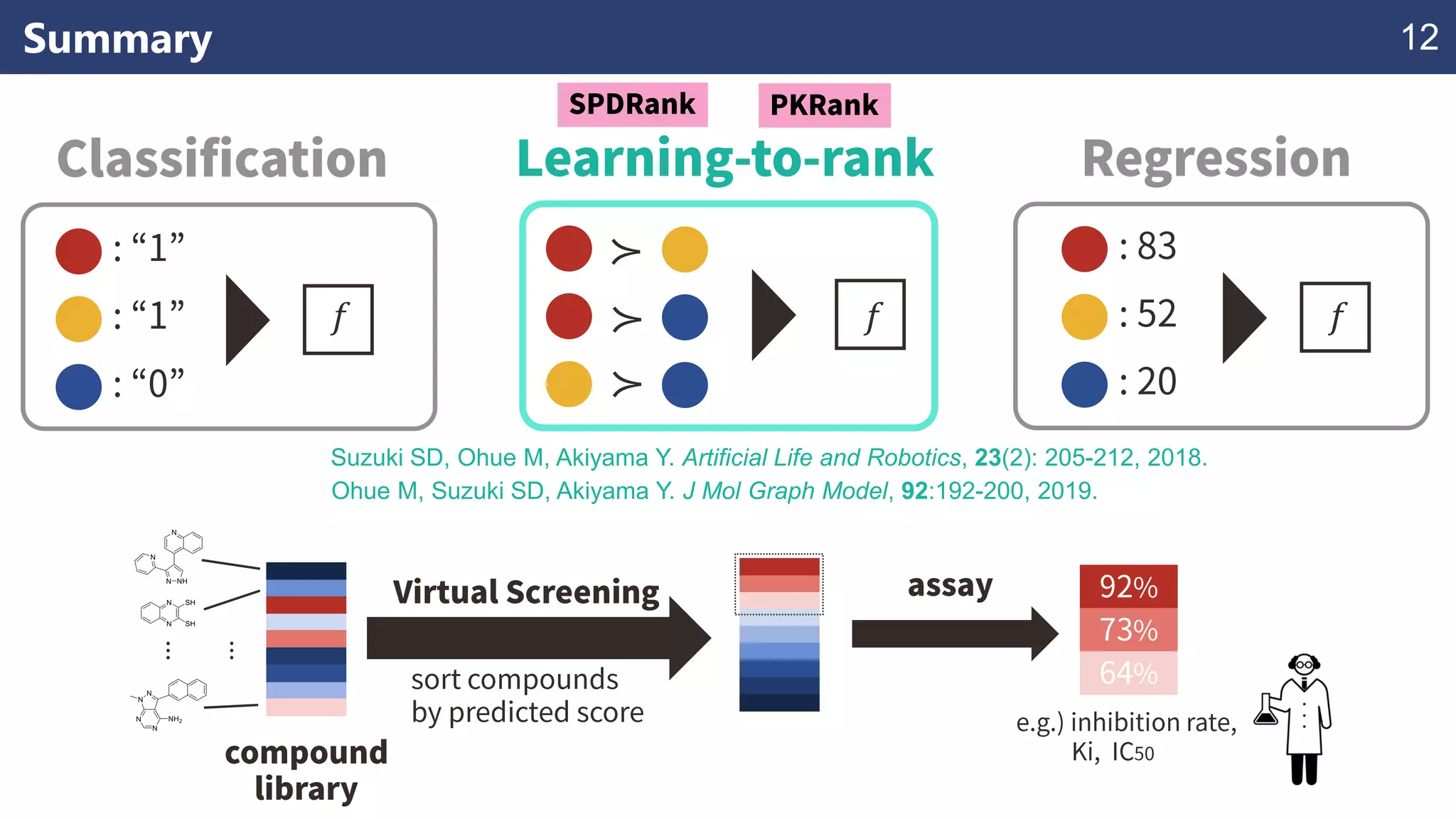
The document discusses the application of learning-to-rank methods for ligand-based virtual screening in drug discovery. It contrasts svm and rank-svm optimization techniques while proposing two new methods for enhancing ligand-based virtual screening results. Additionally, it highlights the integration of diverse experimental data to improve prediction accuracy and minimize irrelevant ordering.