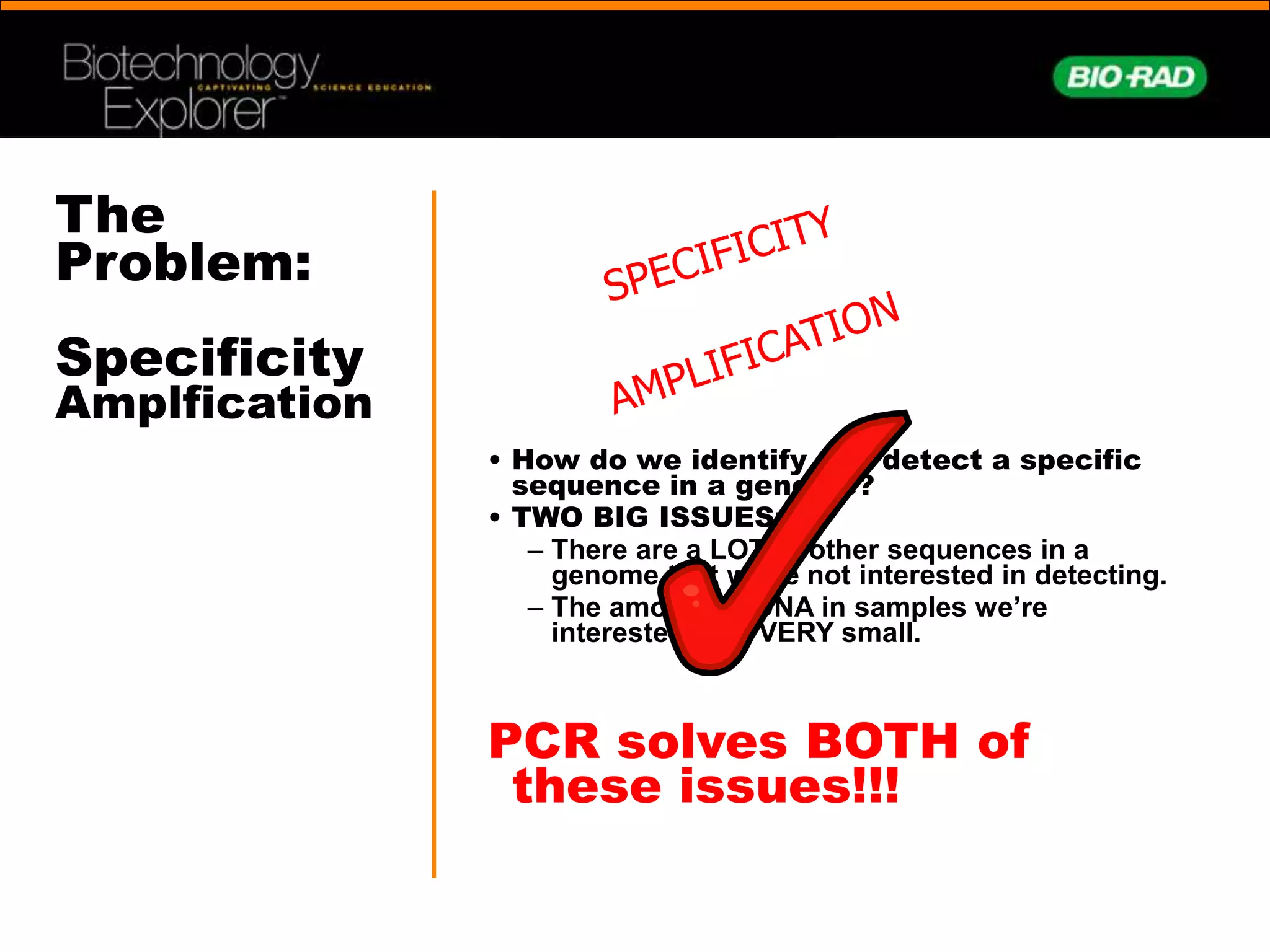
This document provides an overview of the history and development of polymerase chain reaction (PCR). It describes how Kary Mullis invented PCR in 1983, allowing rapid amplification of specific DNA sequences. This enabled DNA to be detected even from very small samples. The document outlines some of the key applications of PCR, such as forensics, disease diagnosis, archaeology, and basic research. It also provides explanations of how PCR works, involving heating and cooling of the DNA sample to facilitate copying of the target sequence through primer-directed polymerase activity.