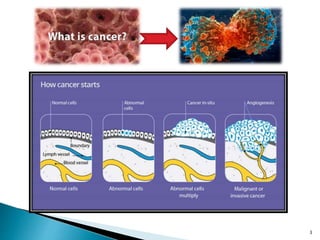
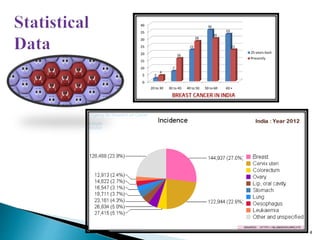
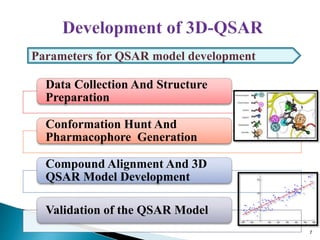
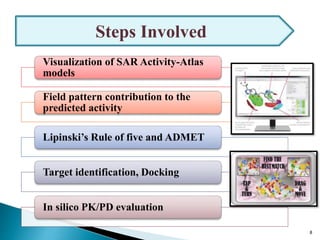
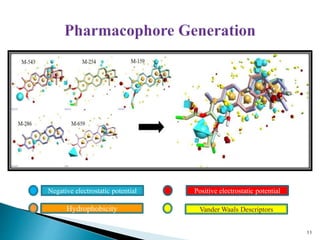
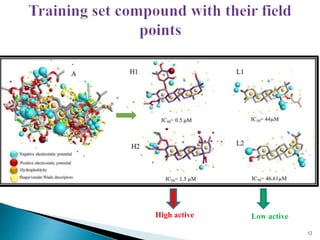
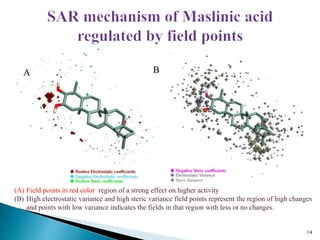
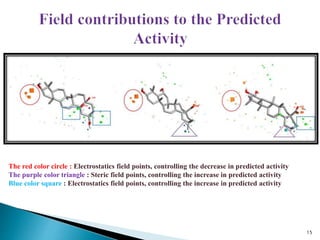
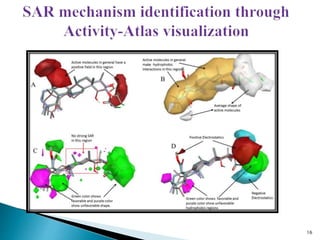
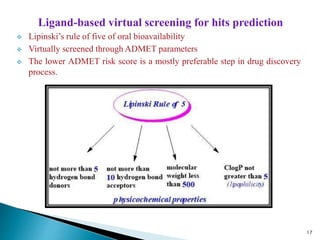
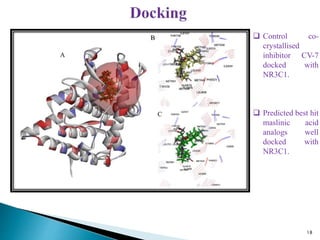
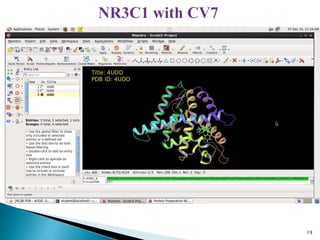
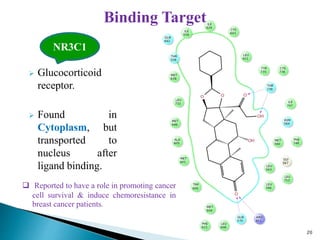
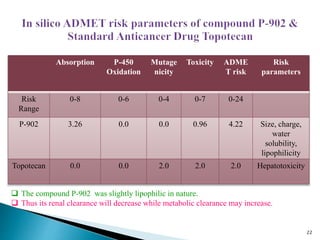
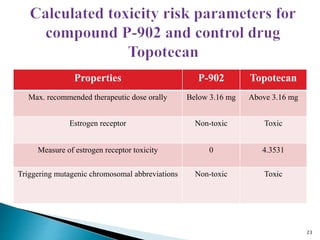
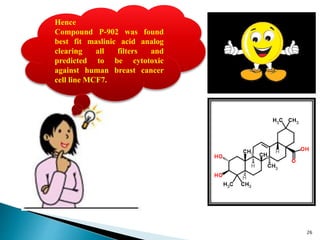
This document presents research on developing a 3D-QSAR model to identify potential anticancer compounds. It describes collecting data on maslinic acid analogs, generating their conformations and pharmacophores, developing a 3D-QSAR model with r2=0.92 and q2=0.75, and validating the model. The model identified compound P-902 as a best fit maslinic acid analog predicted to be cytotoxic against breast cancer cells through ligand-based virtual screening, ADMET analysis, and docking studies targeting the glucocorticoid receptor NR3C1.