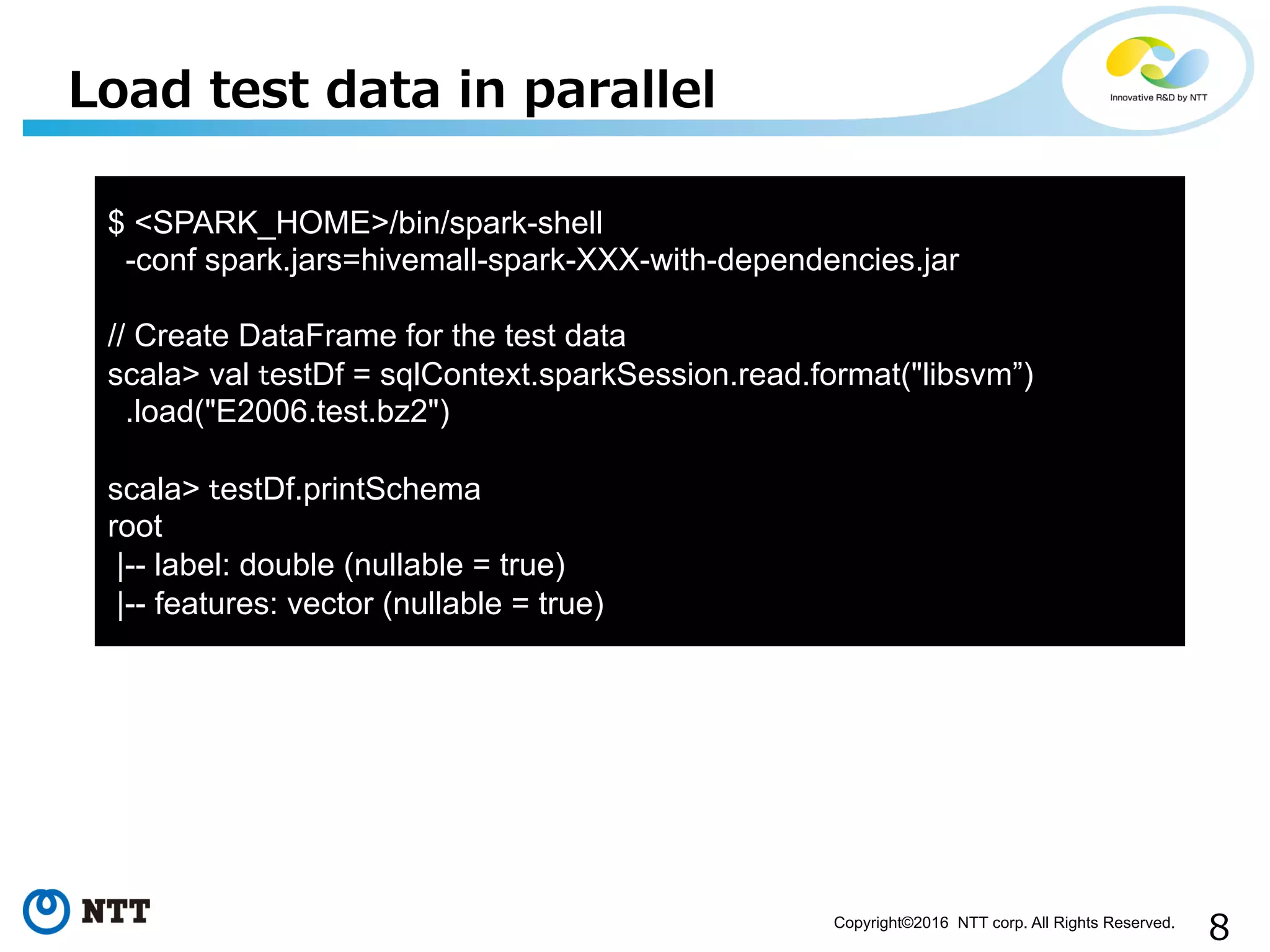
This document discusses integrating XGBoost machine learning with Spark and DataFrames. It provides examples of using XGBoost in Spark to train models on distributed data and make predictions on streaming data in parallel. It also discusses future work, such as using Rabbit for parallel learning, adding support to more platforms like Windows, and integrating with Spark ML pipelines.