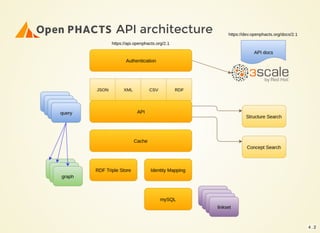
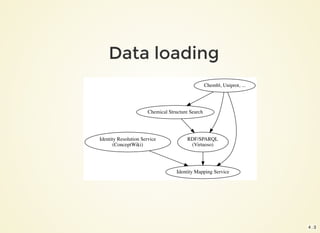
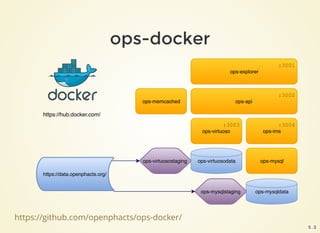
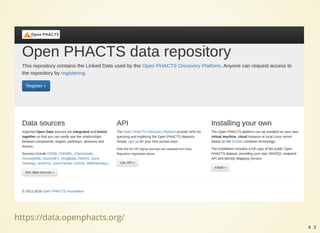
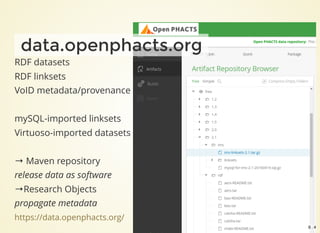
The document outlines the architecture and installation of the Open PHACTS platform, which integrates various pharmacological data resources into a linked and interoperable infrastructure. It covers data integration procedures, API architecture using Docker for microservices, and hardware requirements for installation. The document also discusses customization options for different user types and emphasizes the importance of scalability and reproducibility in bioinformatics workflows.

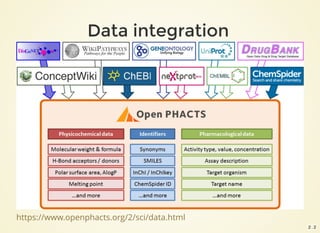

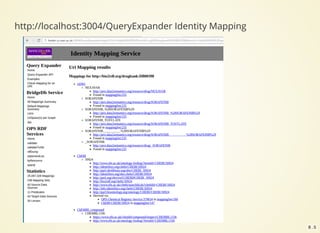
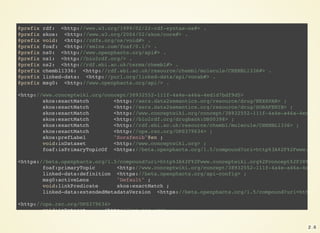
![{
"format": "linked-data-api",
"version": "1.5",
"result": {
"_about": "https://beta.openphacts.org/1.5/compound?uri=http%3A%2F%2Fwww.conceptwiki.org%2Fconce
"definition": "https://beta.openphacts.org/api-config",
"extendedMetadataVersion": "https://beta.openphacts.org/1.5/compound?uri=http%3A%2F%2Fwww.concep
"linkPredicate": "http://www.w3.org/2004/02/skos/core#exactMatch",
"activeLens": "Default",
"primaryTopic": {
"_about": "http://www.conceptwiki.org/concept/38932552-111f-4a4e-a46a-4ed1d7bdf9d5",
"inDataset": "http://www.conceptwiki.org",
"exactMatch": [
{
"_about": "http://bio2rdf.org/drugbank:DB00398",
"description_en": "Sorafenib (rINN), marketed as Nexavar by Bayer, is a drug approved for th
"description": "Sorafenib (rINN), marketed as Nexavar by Bayer, is a drug approved for the t
"drugType_en": [
"investigational",
"approved"
],
"drugType": [
"investigational",
"approved"
],
"genericName_en": "Sorafenib",
"genericName": "Sorafenib",
"metabolism_en": "Sorafenib is metabolized primarily in the liver, undergoing oxidative meta
"metabolism": "Sorafenib is metabolized primarily in the liver, undergoing oxidative metabol
"proteinBinding_en": "99.5% bound to plasma proteins.",
"proteinBinding": "99.5% bound to plasma proteins.",
"toxicity_en": "The highest dose of sorafenib studied clinically is 800 mg twice daily. The
"toxicity": "The highest dose of sorafenib studied clinically is 800 mg twice daily. The adv
"inDataset": "http://www.openphacts.org/bio2rdf/drugbank", 2 . 4](https://image.slidesharecdn.com/2016-07-06-openphacts-docker-160707133429/85/2016-07-06-openphacts-docker-5-320.jpg)
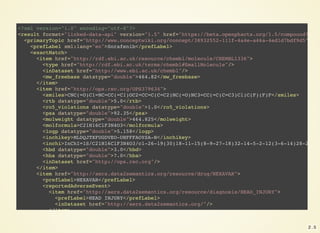







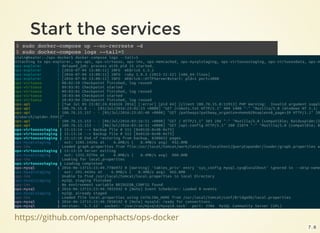
![https://github.com/openphacts/ops-docker
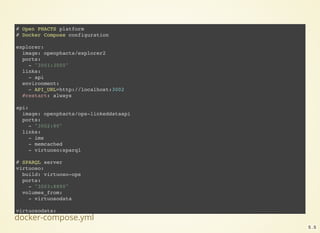
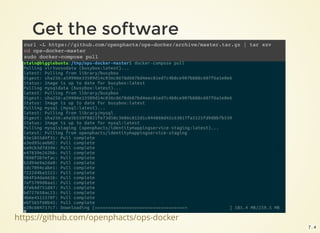
Get the data
$ sudo docker-compose up --no-recreate -d mysqlstaging virtuosostaging
$ sudo docker-compose logs mysqlstaging virtuosostaging
ops-mysqlstaging | mySQL staging finished
ops-mysqlstaging exited with code 0
ops-virtuosostaging | 09:13:35 --> Backup file # 675 [0x3F02-0x74-0x8A]
ops-virtuosostaging | 09:13:36 --> Backup file # 676 [0x3F02-0x74-0x8A]
ops-virtuosostaging | 09:13:37 End of restoring from backup, 6751701 pages
ops-virtuosostaging | 09:13:37 Server exiting
ops-virtuosostaging | Loading completed
ops-virtuosostaging exited with code 0
7 . 5](https://image.slidesharecdn.com/2016-07-06-openphacts-docker-160707133429/85/2016-07-06-openphacts-docker-25-320.jpg)