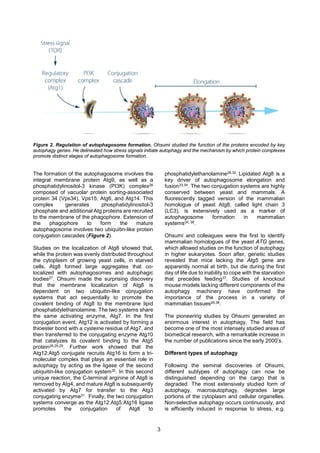
Yoshinori Ohsumi's seminal work in the early 1990s dramatically advanced the understanding of autophagy. Using yeast as a model organism, he identified 15 essential autophagy genes and developed yeast strains that allowed him to discover the first autophagy gene, Atg1. His subsequent cloning and characterization of additional autophagy genes elucidated their protein products and roles in autophagosome formation. This included delineating how stress signals initiate autophagy through the Atg1 kinase complex and phosphatidylinositol 3-kinase complex, and the two ubiquitin-like conjugation systems that promote phagophore extension and autophagosome maturation. Ohsumi