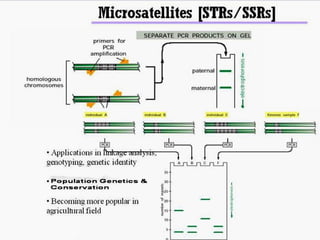
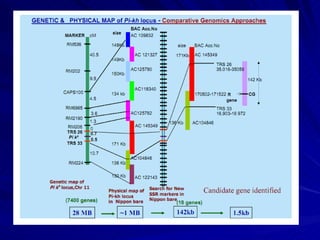
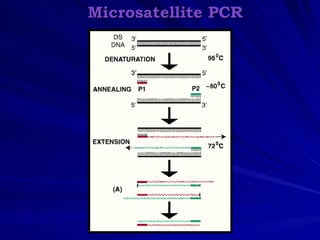
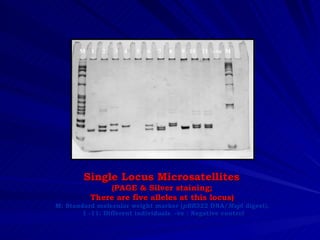
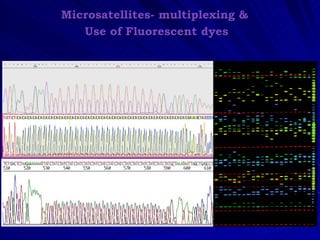
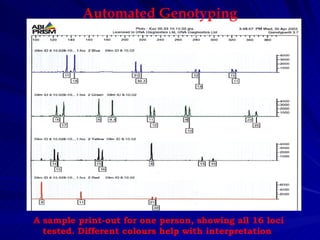
Microsatellites are tandemly repeated DNA sequences with repeat units of 1-6 base pairs. They are highly polymorphic due to variations in the number of repeats between individuals. Microsatellites can be analyzed using PCR and electrophoresis to differentiate alleles and study genetic diversity, population structure, and parentage. A genetic map of microsatellites was constructed for turbot fish to enable future quantitative trait locus identification and evolutionary studies. Microsatellites are a powerful tool for various areas of genetics research.


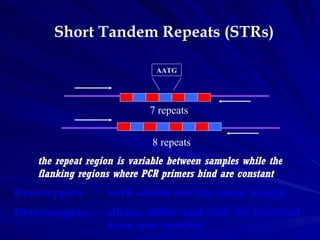



![Microsatellites – Properties
Co-dominant
Inherit in Mendelian Fashion
Polymorphic loci with allele number as high as 14 – 15
per locus
Mostly reported from non-coding region, hence can be
independent of selection
Flanking region is highly conserved in related species
Can be obtained from small amounts of tissues [STR
analysis can be done on less than one billionth of a
gram (a nanogram) of DNA (as in a single flake of
dandruff)]
PAGE separation; silver staining/automated genotyping
Abundant in the eukaryote genome (~103 to 105 loci
dispersed at 7 to 10100 kilobase pair (kb) intervals)](https://image.slidesharecdn.com/microsatellite-120515062853-phpapp01/85/Microsatellite-7-320.jpg)