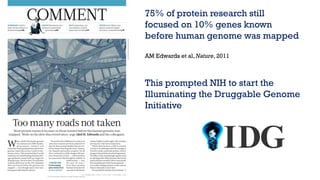
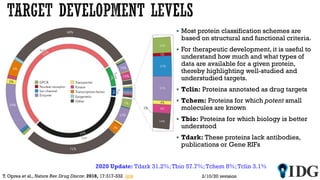
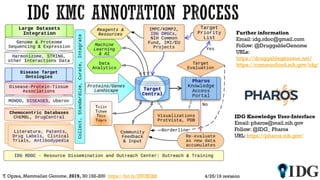
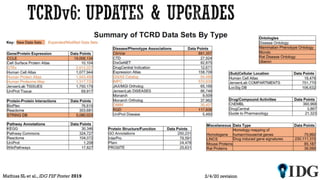
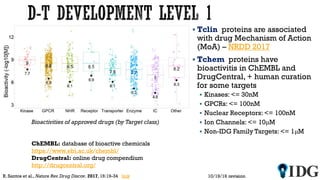
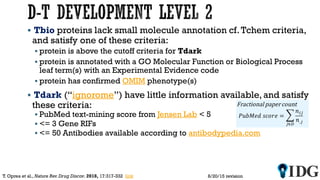
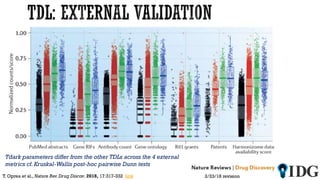
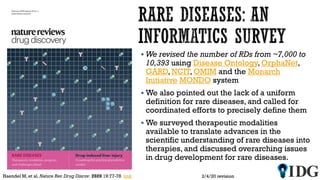
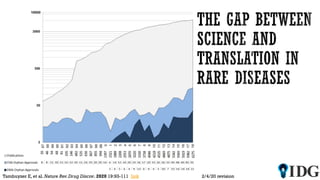
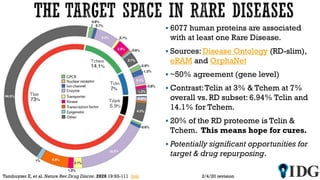
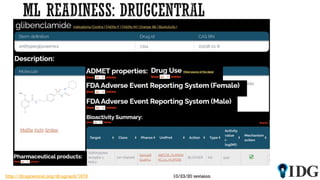
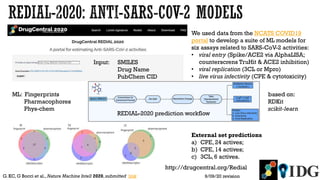
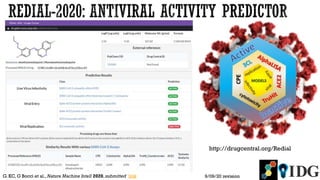
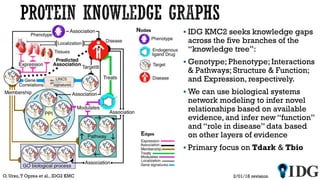
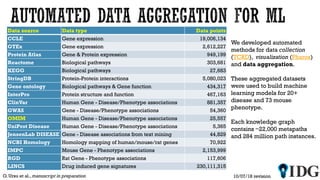
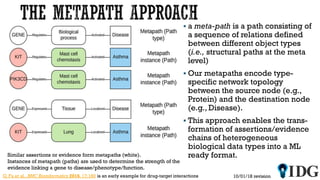
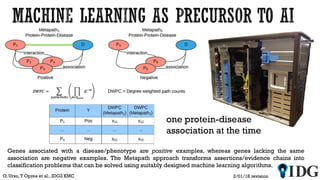
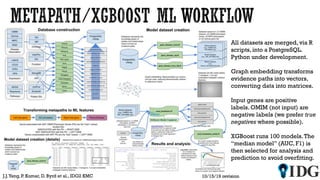
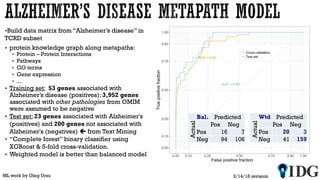
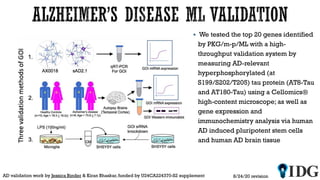
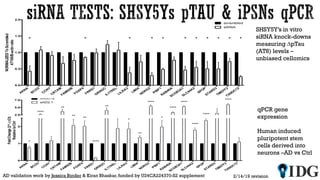
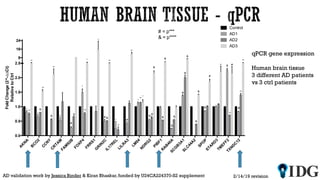
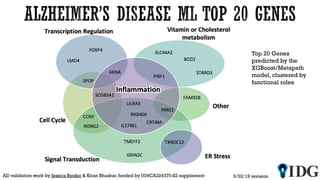
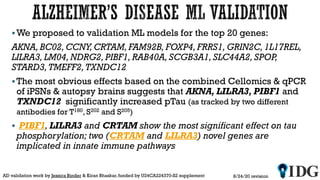
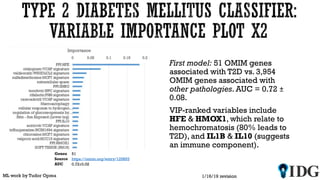
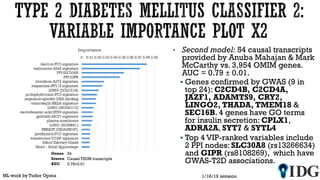
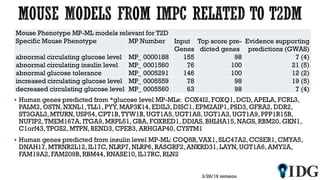
The document discusses the Illuminating the Druggable Genome initiative, which aims to enhance protein-targeting research through informatics, data science, and machine learning. It highlights the classification of proteins associated with drug targets, detailing the available data for various protein categories and the significance of understanding under-studied proteins in therapeutic development. Additionally, the document emphasizes the potential of machine learning in prioritizing clinically relevant protein-disease associations and the aggregation of diverse biological data to inform drug development.