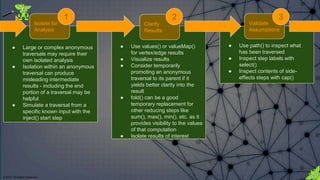
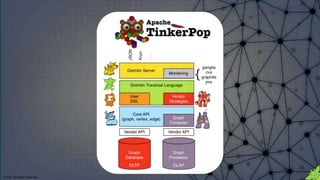
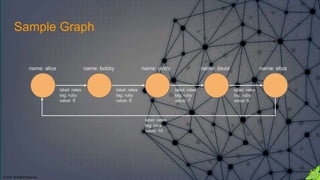
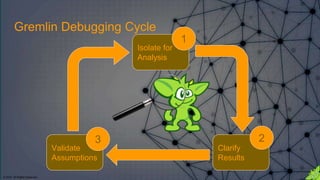
The document discusses the anatomy of Gremlin, a graph traversal language, detailing its components like vertices, edges, and traversal steps. It emphasizes the importance of understanding Gremlin to improve code writing, debugging, and building domain-specific languages. Additionally, it introduces techniques for analyzing and validating Gremlin queries through a debugging cycle.












![© 2018. All Rights Reserved.
gremlin> graph = TinkerGraph.open()
==>tinkergraph[vertices:0 edges:0]
gremlin> g = graph.traversal()
==>graphtraversalsource[tinkergraph[vertices:0 edges:0], standard]
gremlin> g.addV().property('name','alice').as('a').
......1> addV().property('name','bobby').as('b').
......2> addV().property('name','cindy').as('c').
......3> addV().property('name','david').as('d').
......4> addV().property('name','eliza').as('e').
......5> addE('rates').from('a').to('b').property('tag','ruby').property('value',9).
......6> addE('rates').from('b').to('c').property('tag','ruby').property('value',8).
......7> addE('rates').from('c').to('d').property('tag','ruby').property('value',7).
......8> addE('rates').from('d').to('e').property('tag','ruby').property('value',6).
......9> addE('rates').from('e').to('a').property('tag','java').property('value',10).
.....10> iterate()
gremlin> graph
==>tinkergraph[vertices:5 edges:5]](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-18-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack(div).
.....19> by(union(constant(1d),
.....20> tail(local, 1)).sum()).
.....21> sack(mult).
.....22> by(limit(local, 1)).
.....23> sack().sum()).
.....24> cap('a')
==>[alice,[alice,bobby,cindy,david,eliza,alice],[9,8,7,6,10],18.833333333333332]](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-19-320.jpg)

![© 2018. All Rights Reserved.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice'))
==>v[0] Isolate a portion of the larger Gremlin
statement for analysis. The isolated portion
should be only as long as you are capable of
following by just reading the code.
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-22-320.jpg)

![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> valueMap()
==>[name:[alice]]
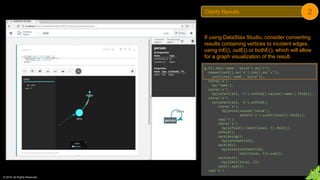
Be clear on the result being returned from the
current portion of Gremlin under review as it
will feed into the following steps that will later
be analyzed.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Clarify Results 2](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-24-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> path().next()
==>v[0]
==>e[10][0-rates->2]
==>v[2]
==>e[11][2-rates->4]
==>v[4]
==>e[12][4-rates->6]
==>v[6]
==>e[13][6-rates->8]
==>v[8]
==>e[14][8-rates->0]
==>v[0]
Validate any assumptions regarding the nature
of the traversal. Consider the traversal path,
the graph schema or anything that might be
hidden by a naive view of the result.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Validate Assumptions 3](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-27-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> select('v')
==>[v[0],v[2],v[4],v[6],v[8],v[0]]
Step labels - created with as()-step - are a
type of side-effect. Have clarity in terms of
what they contain. Use select()-step to extract
their contents.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Validate Assumptions 3](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-28-320.jpg)

![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name')
==>v[0]
Slowly expand the portion of Gremlin being
analyzed to the next point of isolation and
repeat the debugging cycle.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-31-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> cap('a')
==>[alice]
As with step labels, verify the contents of side-
effects. In this case, determine what is being
stored in the "a" side-effect by using cap()-
step.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Examine Side-effects 3](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-33-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold())
==>v[0]
Slowly expand the portion of Gremlin being
analyzed to the next point of isolation and
repeat the debugging cycle.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-34-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> cap('a')
==>[alice,[alice,bobby,cindy,david,eliza,alice]]
As with step labels, verify the contents of side-
effects. In this case, determine what is being
stored in the "a" side-effect by using cap()-
step.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Examine Side-effects 3](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-35-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v')).
......7> cap('a')
==>[alice,[v[0],v[2],v[4],v[6],v[8],v[0]]]
To gain clarity on anonymous traversals,
isolate them for review as part of the
debugging cycle.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-37-320.jpg)
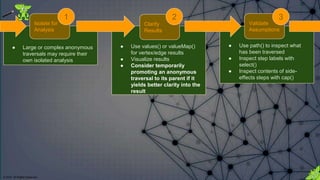
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> select(all, 'v')
==>[v[0],v[2],v[4],v[6],v[8],v[0]]
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> select(all, 'v').
......4> unfold().
......5> values('name')
==>alice
==>bobby
==>cindy
==>david
==>eliza
==>alice
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> select(all, 'v').
......4> unfold().
......5> values('name').
......6> fold()
==>[alice,bobby,cindy,david,eliza,alice]
For analysis consider temporarily promoting
the anonymous traversal to the parent
traversal as it might allow better focus on the
actual result. In this case, the store() side-
effects are not relevant to the specific analysis
of the contents of the by() modulator.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Clarify Results 2A
B
C](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-39-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack(div).
.....19> by(union(constant(1d),
.....20> tail(local, 1)).sum()).
.....21> sack(mult).
.....22> by(limit(local, 1)).
.....23> sack().sum()).
.....24> cap('a')
==>[alice,[alice,bobby,cindy,david,eliza,alice],[9,8,7,6,10],18.833333333333332]](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-40-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold())).
.....15> cap('a').next()
==>alice
==>[alice,bobby,cindy,david,eliza,alice]
==>[9,8,7,6,10]
==>[[9,0],[8,1],[7,2],[6,3],[10,4]]
It may be necessary to isolate anonymous
traversals if they are especially complex.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-41-320.jpg)
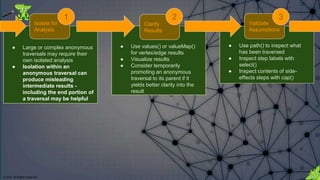
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold()).
.....16> cap('a').next()
==>alice
==>[alice,bobby,cindy,david,eliza,alice]
==>[9,8,7,6,10]
==>[9,0]
Analyzing portions of a traversal can produce
misleading intermediate results that might
leave an incorrect impression as to the overall
intention of the traversal when executed as a
whole.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-43-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d))).
.....18> cap('a').next()
==>alice
==>[alice,bobby,cindy,david,eliza,alice]
==>[9,8,7,6,10]
==>[9,0]
Analyzing portions of a traversal can produce
misleading intermediate results that might
leave an incorrect impression as to the overall
intention of the traversal when executed as a
whole.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-44-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack().sum()).
.....19> cap('a').next()
==>alice
==>[alice,bobby,cindy,david,eliza,alice]
==>[9,8,7,6,10]
==>5.0
Include context from the end of a portion of a
traversal to help ensure the intent of the
overall traversal is consistent.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-45-320.jpg)
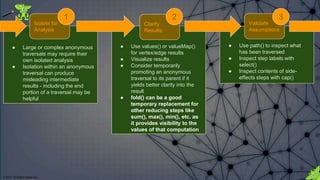
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack().fold()).
.....19> cap('a').next()
==>alice
==>[alice,bobby,cindy,david,eliza,alice]
==>[9,8,7,6,10]
==>[1.0,1.0,1.0,1.0,1.0]
Do not allow the actual steps in the traversal to
prevent visibility into the nature of the
computations that Gremlin is performing.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Clarify Results 2](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-47-320.jpg)
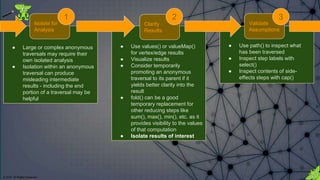
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack(div).
.....19> by(union(constant(1d),
.....20> tail(local, 1)).sum()).
.....21> sack().fold()).
.....22> cap('a').unfold().skip(3).fold().next()
==>[1.0,0.5,0.3333333333333333,0.25,0.2]
Limit output as necessary to focus on the
aspects of the results that are most under
consideration.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Clarify Results 2](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-49-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.inject([9,0],[8,1],[7,2],[6,3],[10,4]).
......1> sack(assign).
......2> by(constant(1d)).
......3> sack(div).
......4> by(union(constant(1d),tail(local,1)).sum()).
......5> sack()
==>1.0
==>0.5
==>0.3333333333333333
==>0.25
==>0.2
gremlin> g.inject([10,4]).
......1> union(constant(1d),tail(local,1))
==>1.0
==>4
gremlin> g.inject([10,4]).
......1> union(constant(1d),tail(local,1)).
......2> sum()
==>5.0
gremlin> 1.0d / g.inject([10,4]).
......1> union(constant(1d),tail(local,1)).
......2> sum().next()
==>0.2
Lost? Reclaim focus by simulating just the
portion of the traversal that is not yet
understood.
g.V().has('name','alice').as('v').
repeat(outE().as('e').inV().as('v')).
until(has('name','alice')).
store('a').
by('name').
store('a').
by(select(all, 'v').unfold().values('name').fold()).
store('a').
by(select(all, 'e').unfold().
store('x').
by(union(values('value'),
select('x').count(local)).fold()).
cap('x').
store('a').
by(unfold().limit(local, 1).fold()).
unfold().
sack(assign).
by(constant(1d)).
sack(div).
by(union(constant(1d),
tail(local, 1)).sum()).
sack(mult).
by(limit(local, 1)).
sack().sum()).
cap('a')
Isolate for Analysis 1
B
A
C
D](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-51-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack(div).
.....19> by(union(constant(1d),
.....20> tail(local, 1)).sum()).
.....21> sack(mult).
.....22> by(limit(local, 1)).
.....23> sack().sum()).
.....24> cap('a')
==>[alice,[alice,bobby,cindy,david,eliza,alice],[9,8,7,6,10],18.833333333333332]](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-52-320.jpg)
![© 2018. All Rights Reserved.
gremlin> g.V().has('name','alice').as('v').
......1> repeat(outE().as('e').inV().as('v')).
......2> until(has('name','alice')).
......3> store('a').
......4> by('name').
......5> store('a').
......6> by(select(all, 'v').unfold().values('name').fold()).
......7> store('a').
......8> by(select(all, 'e').unfold().
......9> store('x').
.....10> by(union(values('value'),
.....11> select('x').count(local)).fold()).
.....12> cap('x').
.....13> store('a').
.....14> by(unfold().limit(local, 1).fold()).
.....15> unfold().
.....16> sack(assign).
.....17> by(constant(1d)).
.....18> sack(div).
.....19> by(union(constant(1d),
.....20> tail(local, 1)).sum()).
.....21> sack(mult).
.....22> by(limit(local, 1)).
.....23> sack().sum()).
.....24> cap('a')
==>[alice,[alice,bobby,cindy,david,eliza,alice],[9,8,7,6,10],18.833333333333332]
Overriding traversers with by()-
modulators
Creating indexed values
Using sack() for computations
Result construction with side-effects
Methods of cycle-detection
Traversal patterns identified as a result
of Gremlin Debugging Cycle
Step label access and collection
manipulation](https://image.slidesharecdn.com/gremlins-anatomy-180223113818/85/Gremlin-s-Anatomy-53-320.jpg)