Eukaryotic Transcription
- 1. Dr. Pawan Kumar Kanaujia Assistant Professor Molecular Biology (Theory) Eukaryotic Transcription
- 6. TRANSCRIPTION IN EUKARYOTES Whereas bacteria have only one RNA polymerase, all eukaryotes have at least three different ones (Pol I, II,and III; and plants have a Pol IV & a Pol V). In addition, whereas bacteria require only one additional initiation factor (σ), several initiation factors are required for efficient and promoter-specific initiation in eukaryotes. These are called the general transcription factors (GTFs). The Subunits of RNA Polymerases
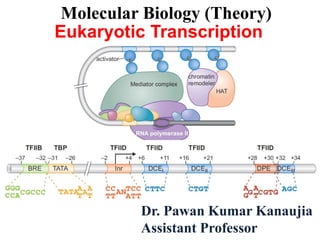
- 7. RNA Polymerase II Core Promoters Are Made Up of Combinations of Different Classes of Sequence Element The eukaryotic core promoter refers to the minimal set of sequence Elements required for accurate transcription initiation by the Pol II machinery. A core promoter is typically ~40–60 nucleotides long, extending either upstream or downstream from the transcription start site. These are the TFIIB recognition element (BRE), the TATA element (or box), the initiator (Inr), and the downstream promoter elements (known as DPE, DCE, and MTE). Typically, a promoter includes some subset of these elements. Thus, for example, promoters typically have either a TATA element or a DPE element, not both. Often, a TATA-containing promoter also contains a DCE. The Inr is the most common element, found in combination with both TATA and DPEs.
- 8. Beyond—and typically upstream of—the core promoter, there are other sequence elements required for accurate and efficient transcription in vivo. Together, these elements constitute the regulatory sequences and can be grouped into various categories, reflecting their location, and the organism. These elements include promoter proximal elements, upstream activator sequences (UASs), enhancers, and a series of other elements called silencers, boundary elements, and insulators. All of these DNA elements bind regulatory proteins (activators and repressors)
- 9. Pol II core promoter Fig 1: The figure shows the positions of various DNA elements relative to the transcription start site (indicated by the arrow above the DNA). These elements, described in the text, are as follows: (BRE) TFIIB recognition element; (TATA) TATA box; (Inr) initiator element; (DPE) downstream promoter element; and (DCE) downstream core element. Another element, MTE (motif ten element), described in the text, is not shown in this figure but is located just upstream of the DPE. Also shown are the consensus sequences for each element and (above) the name of the general transcription factor that recognizes each element.
- 10. RNA Polymerase II Forms a Preinitiation Complex with General Transcription Factors at the Promoter The general transcription factors collectively perform the functions as Performed by σ in bacterial transcription. Thus, the general transcription factors help polymerase bind to the promoter and melt the DNA (comparable to the transition from the closed to open complex in the bacterial case). They also help polymerase escape from the promoter and embark on the elongation phase. The complete set of general transcription factors and polymerase, bound together at the promoter and poised (rediness for action) for initiation, is called the preinitiation complex. Pol II promoters contain a so-called TATA element (some -30 bp upstream of the transcription start site), where preinitiation complex formation begins. The TATA element is recognized by the general transcription factor called TFIID. The component of TFIID that binds to the TATA DNA sequence is called TBP (TATA-binding protein).
- 11. The other subunits in this complex are called TAFs (TBP-associated factors). TAFs recognize other core promoter elements such as the Inr, DPE, and DCE, although the strongest binding is between TBP and TATA. Thus, TFIID is a critical factor in promoter recognition and preinitiation complex establishment. TBP–DNA complex provides a platform to recruit other general transcription factors and polymerase itself to the promoter. In vitro, these proteins assemble at the promoter in the following order (Fig 1): TFIIA, TFIIB, TFIIF together with polymerase, and then TFIIE and TFIIH. Formation of the preinitiation complex containing these components is followed by promoter melting. In eukaryotes requires hydrolysis of ATP and is mediated by TFIIH.
- 12. Fig 2: The stepwise assembly of the Pol II preinitiation complex is shown here and described in detail in the previous or above slide. Once assembled at the promoter, Pol II leaves the preinitiation complex upon addition of the nucleotide precursors required for RNA synthesis and after phosphorylation of serine resides within the enzyme’s “tail”. The tail contains multiple repeats of the heptapeptide sequence: Tyr-Ser-Pro-Thr-Ser- Pro-Ser Transcription initiation by RNA Pol II
- 13. Promoter Escape Requires Phosphorylation of the Polymerase “Tail” During abortive initiation, the polymerase synthesizes a series of short transcripts. In eukaryotes, promoter escape involves two steps not seen in bacteria: one is ATP hydrolysis (in addition to the earlier ATP hydrolysis needed for DNA melting), and the other is phosphorylation of the polymerase. The large subunit of Pol II has a carboxy-terminal domain (CTD), which is referred to as the “tail” (see Fig 2). The CTD contains a series of repeats of the heptapeptide sequence: Tyr-Ser-Pro-Thr-Ser-Pro-Ser. There are 27 of these repeats in the yeast Pol II CTD, 32 in the worm, 45 in the fly Drosophila and 52 in humans. Indeed, the number of repeats seems to correlate with the complexity of the genome. Each repeat contains sites for phosphorylation by specific kinases, including one that is a subunit of TFIIH. When Pol II recruited to the promoter initially unphosphorylated tail, but the species found in the elongation complex bears multiple phosphoryl groups on its tail. Phosphorylation state of the CTD of Pol II controls subsequent steps— elongation and even processing of the RNA—as well.
- 14. The Other General Transcription Factors Also Have Specific Roles in Initiation Table: The General Transcription Factors of RNA Polymerase II
- 15. TAFs :- TBP is associated with about 10 TAFs. Two of the TAFs bind DNA elements at the promoter, for example, the initiator element (Inr) and the downstream promoter elements (Fig 1). TFIIB:- This protein, a single polypeptide chain, enters the preinitiation complex after TBP (Fig 2). The crystal structure of the ternary complex of TFIIB–TBP–DNA shows specific TFIIB–TBP and TFIIB–DNA contacts (Fig 4). These include base-specific interactions with the major groove upstream (to the BRE) (Fig 1) and the minor groove downstream of the TATA element. TFIIF :- This two-subunit (in humans) factor associates with Pol II and is recruited to the promoter together with that enzyme (and other factors). Binding of Pol II–TFIIF stabilizes the DNA–TBP–TFIIB complex and is required before TFIIE and TFIIH are recruited to the preinitiation complex (Fig2). TFIIE and TFIIH:- TFIIE, which, like TFIIF, consists of two subunits, binds next and has roles in the recruitment and regulation of TFIIH. TFIIH controls the ATP-dependent transition of the preinitiation complex to the open complex.
- 16. In Vivo, Transcription Initiation Requires Additional Proteins, Including the Mediator Complex One reason for these additional requirements is that the DNA template in vivo is packaged into chromatin. This condition complicates binding to the promoter of polymerase and its associated factors. Transcriptional regulatory proteins called activators help recruit polymerase to the promoter, stabilizing its binding there. This recruitment is mediated through interactions between DNA-bound activators, chromatin-modifying and -remodeling factors, and parts of the transcription machinery. Mediator is associated with the basic transcription machinery, most likely touching the CTD “tail” of the large polymerase subunit through one surface, while presenting other surfaces for interaction with DNA-bound activators.
- 17. Assembly of the preinitiation complex in the presence of Mediator, nucleosome modifiers and remodelers, and transcriptional activators Fig 5: Tanscriptional activators bound to sites near the gene recruit nucleosome-modifying and –remodeling complexes and the Mediator complex, which together help form the preinitiation complex.
- 18. A New Set of Factors Stimulates Pol II Elongation and RNA Proofreading Once polymerase has escaped the promoter and initiated transcription, it shifts into the elongation phase, as we have discussed. This transition involves the Pol II enzyme shedding most of its initiation factors—for example, the general transcription factors and Mediator. In their place, another set of factors is recruited. Some of these (such as TFIIS and SPT5) are elongation factors (i.e., factors that stimulate elongation). In this case, however, the factors favor the phosphorylated form of the CTD. Thus, phosphorylation of the CTD leads to an exchange of initiation factors for those factors required for elongation and RNA processing. Together, these features allow the tail to bind several components of the elongation and processing machinery and deliver them to the emerging RNA. Various proteins are thought to stimulate elongation by Pol II. One of these, the kinase P-TEFb, is recruited to polymerase by transcriptional activators.
- 19. Once bound to Pol II, this protein phosphorylates the serine residue at position 2 of the CTD repeats. That phosphorylation event correlates with elongation (Fig 6) In addition, P-TEFb phosphorylates and thereby activates another protein, called SPT5, itself an elongation factor. Finally, TAT-SF1, yet another elongation factor, is recruited by P-TEFb.
- 20. Fig 6:-RNA processing enzymes are recruited by the CTD tail of polymerase
- 21. Fig 6: (a) Various factors involved in RNA processing recruited by the CTD tail of polymerase. Different factors are recruited depending on the phosphorylation state of the tail. Those factors are then transferred to the RNA as they are needed (see next section in text). (b) A schematic of the tail, with the sequence of one copy of the heptapeptide repeat shown in the top line. The positions of serine residues that get phosphorylated are indicated in lines 2 and 3. Phosphorylation of serine at position 5 is seen upon promoter escape and is associated with recruitment of capping factors, where as phosphorylation of serine at position 2 is seen during elongation and is associated with recruitment of splicing factors. Recruitment of factors involved in elongation of transcription and in RNA processing overlaps. Thus, elongation factor hSPT5 is recruited to the tail phosphorylated on Ser-5.
- 22. Elongating RNA Polymerase Must Deal with Histones in Its Path As with initiation of transcription, elongation also takes place in the presence of histones, because the DNA template is incorporated into nucleosomes. Factor called FACT (facilitates chromatin transcription) was identified in human makes transcription on chromatin templates much more efficient. FACT is a heterodimer of two well conserved proteins, Spt16 and SSRP1. Nucleosomes are octomers, made up of H2A, H2B, H3,and H4 histone subunits and DNA. These histones are arranged in two modules: the H2A.H2B dimers and the H3.H4 tetramer. Ahead of a transcribing RNA polymerase, FACT removes one H2A.H2B dimer. This allows polymerase to pass that nucleosome . FACT also has histone chaperone activity, which allows it to restore the H2A.H2B dimer to the histone hexamer immediately behind the Processing polymerase. In this way, FACT allows polymerase to elongate and at the same time maintains the integrity of the chromatin through which the enzyme is transcribing.
- 23. A model for FACT-aided elongation through nucleosomes Fig 8:- Step 1- FACT, shown as the heterodimer of Spt16 and SSRP1, is able to dismantle nucleosomes ahead of the transcribing RNA polymerase and reassemble them behind. Step 2- Specifically, it removes the H2A.H2B dimer. SPT6 binds histone H3 and is believed to aid in nucleosome reassembly.
- 24. Elongating Polymerase Is Associated with a New Set of Protein Factors Required for Various Types of RNA Processing These processing events include capping of the 5’ end of the RNA, splicing, and polyadenylation of the 3’ end of the RNA. The most complicated of these is splicing—the process whereby non-coding introns are removed from RNA to generate the mature mRNA. Here we consider the other two processes—capping and polyadenylating the transcript. In one case, for example, an elongation factor mentioned above (SPT5) also helps to recruit the 5’-capping enzyme to the CTD tail of polymerase (phosphorylated at serine position 5) (Fig 6b).The hSPT5 stimulates the 5’-capping enzyme activity. In another case, elongation factor TAT-SF1 recruits components of the splicing machinery to polymerase with a Ser-2 phosphorylated tail (Fig 6b). Thus, elongation, termination of transcription, and RNA processing are interconnected, presumably to ensure their proper coordination.
- 25. The 5’ cap is created in three enzymatic steps, as detailed in Fig 9 and described in detail in the legend. In the first step, a phosphate group is removed from the 5’ end of the transcript. Then, in the second step, the GMP moiety is added. In the final step, that nucleotide is modified by the addition of a methyl group. The RNA is capped as soon as it emerges from the RNA-exit channel of polymerase. This happens when the transcription cycle has progressed only as far as the transition from the initiation to elongation phases. After capping, dephosphorylation of Ser-5 within the tail repeats may be responsible for dissociation of the capping machinery, and further phosphorylation (this time of Ser-2 within the tail repeats) causes recruitment of the machinery needed for RNA splicing (Fig 6b)
- 26. Fig 9:- In the first step, the ϒ-phosphate at the 5’ end of the RNA is removed by an enzyme called RNA triphosphatase (the initiating nucleotide of a transcript initially retains its α-, β-, and ϒ phosphates). In the next step, the enzyme guanylyltransferase adds a GMP moiety to the resulting terminal β-phosphatase. This is a two-step process: first, an enzyme–GMP complex is generated from GTP with release of the β- and ϒ -phosphates of that GTP, and then the GMP from the enzyme is transferred to the β-phosphate of the 5’ end of the RNA. Once this linkage is made, the newly added guanine and the purine at the original 5’ end of the mRNA are further modified by the addition of methyl groups by methyltransferase. The resulting 5’ cap structure subsequently recruits the ribosome to the mRNA for translation to begin. The structure and formation of the 5’RNA cap
- 27. The final RNA processing event, polyadenylation of the 3’ end of the mRNA, is intimately linked with the termination of transcription (Fig 10). Just as with capping and splicing, the polymerase CTD tail is involved in recruiting some of the enzymes necessary for polyadenylation (Fig 6). Once polymerase has reached the end of a gene, it encounters specific sequences that, after being transcribed into RNA, trigger the transfer of the polyadenylation enzymes to that RNA, leading to four events: Cleavage of the message; addition of many adenine residues to its 3’ end; Degradation of the RNA remaining associated with RNA pol by a 5’-to-3’ ribonuclease; and, subsequently, termination of transcription. This series of events unfolds as follows.
- 28. Two protein complexes are carried by the CTD of polymerase as it approaches the end of the gene: CPSF (cleavage and polyadenylation specificity factor)and CSTF (cleavage stimulation factor). The sequences that, once transcribed into RNA, trigger transfer of these factors to the RNA are called poly-A signals, and their operation is shown in Fig 10. Once CPSF and CSTF are bound to the RNA, other proteins are recruited as well, leading initially to RNA cleavage and then polyadenylation. Polyadenylation is mediated by an enzyme called poly-A polymerase, which adds approximately 200 adenines to the RNA’s 3’ end produced by the cleavage. This enzyme uses ATP as a precursor and adds the nucleotides using the same chemistry as RNA polymerase. But it does so without a template. Thus, the long tail of As is found in the RNA but not the DNA. The mature mRNA is then transported from the nucleus.
- 29. Polyadenylation and termination Fig 10:-The various steps in this Process are described in the above text.
- 30. Transcription Termination Is Linked to RNA Destruction by a Highly Processive RNase Polyadenylation is linked to termination, although exactly how is still not quite clear. Recently, however, an enzyme that degrades the second RNA as it emerges from the polymerase has been identified, and this enzyme may itself trigger termination. This is called the torpedo model of termination (Fig 11a). The free end of the second RNA is uncapped and thus can be distinguished from genuine transcripts. This new RNA is recognized by an RNase called, in yeast, Rat1 (in humans, Xrn2) that is loaded onto the end of the RNA by another protein (Rtt103) that binds the CTD of RNA pol. The Rat1 enzyme is very processive and quickly degrades the RNA in a 5’-to-3’ direction, until it catches up to the still-transcribing polymerase from which the RNA is being spewed. Although the torpedo model for termination is now the favored one, there is an alternative called the allosteric model (Fig 11b). According to this model, termination depends on a conformational change in the elongating polymerase that reduces the processivity of the enzyme leading to spontaneous termination soon afterward.
- 31. Fig 11:- Models of termination: torpedo and allosteric
- 32. Fig 11:- there are two proposed models for how transcription by eukaryotic RNA Pol II terminates after transcribing a gene. In the figure, the poly-A site is marked by the light green stretch in the DNA and is located just downstream from the gene. It is also light green in the transcript. (The dotted green line) Degraded transcript. (a) In the torpedo model, RNA transcribed downstream from the poly-A site is attacked by the 5’-to-3’ RNase (the torpedo), which is loaded onto this transcript from polymerase itself. When this exonuclease catches up with polymerase, it triggers dissociation from the DNA template and termination of transcription. (b) In the allosteric model, the polymerase is highly processive within the gene, and then, once the poly-A signal is passed, becomes less processive. This alteration could be due to a modification or a conformational change. Even in the allosteric model, the second RNA would be degraded by the RNase, but that would not be the cause of termination. In this case, RNA degradation is not shown in the figure to emphasize the different mechanisms of termination in these two models.
- 33. Thank you