Plant_Transcription_factors
- 4. RNA
- 5. Factor
- 6. Factor - Something which has an effect on the result
- 7. X factor
- 8. 2 x = 8 x = 8/2 x = 4
- 9. Nuclear Transcription Factor Y Transcription Initiation Factor
- 10. General TF / Specific TF
- 11. General Transcriptional factors 5 I 3 I
- 12. Pol II General transcription factors TATAAA -30 +1 Transcription Start site TF II D TBP TF II A TF II B TF II F TF II E TF II H • TF II D - TBP(Core promoter recognition and recruiting TF II B • TF II A - Stabilising TBP • TF II B - Moving of pol II and TF II F to site and determination of transcriptional initiation point by pol II • TF II F - Fixation of pol II and releasing non specific interaction between pol II and DNA • TF II E - Moving of TF II H , regulation of activity of TF II H helices, ATPase, kinase • TF II H - Conversion of Double stranded DNA into Single stranded
- 14. Specific Transcriptional factors 5 I 3 I
- 15. Transcription factors : Total number of genes 4.7% 3.6 % 5 - 10 %
- 16. Classification of Transcription Factors
- 19. Classification of Transcription Factors*
- 20. bZIP Transcription factor family
- 22. Light signalling Abiotic stress response Pathogen defence Seed maturation Flower development
- 24. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group A Seven members of group A have been studied (AtbZIP39/ABI5, AtbZIP36/ABF2/AREB1, AtbZIP38/ABF4/AREB2, AtbZIP66/AREB3, AtbZIP40/GBF4, AtbZIP35/ABF1 and AtbZIP37/ABF3) ABA / Stress signaling
- 25. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group B Group-B bZIP proteins include AtbZIP17, AtbZIP28, and AtbZIP49 Endoplasmic reticulum (ER) stress responses
- 26. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group C Members of this group share structural features with a well characterized family of plant bZIPs that includes maize Opaque2 and parsley (Petroselinum crispum) CPFR2 Seed storage protein/ Pathgen response ?
- 27. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group D Group D genes participate in two different processes: defence against pathogens and development. Two group D genes are involved in developmental processes: AtbZIP46 /Perianthia controls floral organ number in Arabidopsis and Liguleless2 establishes the blade- sheath boundary during maize leaf development AtbZIP57/OBF4/TGA4 interacts with AtEBP, which binds the ethylene response element present in many PR gene promoters might thus be involved in integrating different systemic signals (salicylic acid and ethylene) at the PR promoter level in response to pathogen infection.
- 28. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group E/F No functional data.
- 29. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group G The group G GBF genes from Arabidopsis and their parsley homologues CPRF1, CPRF3, CPRF4a and CPRF5 have been mainly linked to ultraviolet and blue light signal transduction and to the regulation of light-responsive promoters
- 30. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group H Group H has only two members (AtbZIP56/HY5 and AtbZIP64). HY5’s role in promoting photomorphogenesis
- 31. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group I Studies of group I genes from several species indicate that they might play a role in vascular development RSG gene from tobacco is specifically expressed in the phloem and activates the GA3 gene of the gibberellin biosynthesis pathway
- 32. 75 Putative genes Clustered into 10 groups A, B, C, D, E, F, G, H, I, S Based sequence similarities of their base region Summary Group S Group S is the largest bZIP group in Arabidopsis but only ATBZIP11/ATB2 has been analysed in detail Transcription of this gene is upregulated by light, in carbohydrate-consuming (i.e. sink) tissue and in the vascular system
- 33. bHLH Transcription factor family
- 35. bHLH Tfs contain the bHLH domain of approximately 60 amino acids, with two functionally distinctive regions, the basic region and the HLH region. The 15 amino acid basic region at the N-terminus of the bHlH domain functions as a DNA- binding motif. The HLH region contains two amphipathic alpha-helices with a linking loop of variable length to form homo or heterodimmers Some bHLH proteins bind to sequences containing a consensus core element called the E-box (5I- CANNTG-3I), with the G-box (5I-CACGTG-3I) being the most common form
- 36. In Arabidopsis 162 bHLH-coding genes have been identified (Bailey et al., 2008). In Rice167 bHLH-coding genes have been identified (Li et al., 2006). The members of the bHLH Tf family in both Arabidopsis and rice are divided into two major groups that contain a canonical bHLH or lack the basic region required for DNA binding. (Li et al., 2006). They are further classified into 25 subfamilies (Li et al., 2006).
- 37. Phytochrome Interacting Factors, (PIFs) which may directly bind to the photoactivated phytochromes. Members of the PIF family have been shown to control light-regulated gene expression directly and indirectly. PIF1, PIF3, PIF4 and PIF5 are degraded in response to light signals, and physical interaction of PIF3 with phytochromes is necessary for the light-induced phosphorylation
- 38. The bHLH Tf MYC2 has been described as a master regulator of the crosstalk between the signaling pathways of JA and those of other phytohormones such as abscisic acid (ABA), salicylic acid (SA), gibberellins (GAs), auxins. Seed coat differentiation, and trichome/root hair formation (MYC1, GL3, EGL3, TT8) (GLABRA3 (GL3), ENHANCER of GLABRA3 (EGL3), TRANSPARENT TESTA8 (TT8)), .
- 39. MYB Transcription factor family
- 41. MYB proteins integrate to a superfamily of Tf. This has the largest number of members of any Arabidopsis Tf family: 197 members with a highly conserved DBD known as the MYB domain This domain generally consists of up to four imperfect amino acid sequence repeats (R) of about 52 amino acids, each forming a helix–turn–helix structure that intercalates in the major groove of the DNA. MYB proteins can be divided into different classes depending on the number of MYB repeats (one to four). I,e R1R2, R1R3, R2R3, R1R2R3 MYB repeats In plants, the MYB family has selectively expanded, particularly through the large family of R2R3-MYB (Dubos et al., 2010). In the Arabidopsis genome, 138 are R2R3-MYB, 5 are R1R2R3- MYB, 52 are MYB- related, and 2 are atypical MYB genes (Yanhui et al., 2006; Katiyar et al., 2012).
- 42. Combinatorial interactions among bHLH Tfs and MYB Tfs are reported to play a key role in flavonoid biosynthesis in plants The maize C1 bHLH protein interacts with the MYB R protein to activate maize flavonoid pathways. Arabidopsis GL3 and EGL3 interact with MYB factors GLABRA1 (Gl1) or WEREWOLF (WER) to form trichomes and root hairs.
- 44. HSF (Heat stress transcription factors)
- 45. Heat stress transcription factors (HSFs) mediate the rapid accumulation of heat shock proteins (HSPs) in response to both heat stress and many chemical stressors HSF recognize conserved binding motifs, so-called heat stress elements (HSE: 5I-AGAAnnTTCT-3I) that exist in the promoters of Hsp genes HSFs have a modular structure with a DBD and an oligomerization domain (OD). In addition, they contain a nuclear localization signal (NLS), a nuclear export signal (NES), and an activator motif (AHA motif). Plant HSFs are classified into three classes, A, B, and C, based on the peculiarities of their ODs. In the Arabidopsis genome, among 21 HSFs,15 belong to Class A, 5 to Class B, and 1 to Class C
- 46. Here functional studies show that HsfA1a has a unique function as master regulator of acquired thermotolerance and trigger of the Hs response and that later on, by interaction with HsfA2 and B1 in a functional triad, affects different aspects of Hs response and recovery
- 47. HsfA2 is the most highly induced HSF in stressed plants and plays a role in thermotolerance and a broader role for expression of general stress-related nonchaperone-encoding genes like APX2 (ASCORBATE PEROXIDASE2)
- 48. Plant specific Transcription factors
- 49. WRKY Transcription factor family
- 50. WRKY ?
- 51. WRKYGQK Tryptophan Arginine Lysine Tyrosine Glycine Glutamine Lysine
- 52. The WRKY Tf family is one of the best studied plant specific Tf families and comprises 74 members in Arabidopsis. The WRKY protein family owes its name to the highly conserved 60 amino acid long WRKY domain, which contains a conserved amino acid sequence motif WRKYGQK at the N-terminus and a novel zinc-finger-like motif at the C-terminus. These two motifs are vital for binding to the consensus cis-acting element termed the W-box (TTGACT/C). Based on both the number of WRKY domains and features of the the zinc- finger motif, WRKY proteins are categorized into three subfamilies: Group I has two WRKY domains, Group II has one WRKY domain with the same Cys2–His2 zinc-finger motif (Group II WRKYs are further divided into a–e based on additional conserved motifs outside the WRKY domain. ) Group III has one WRKY domain containing a different Cys2–His2 zinc-finger motif.
- 53. AtWRKY52/ RRs1, a member of Group III that contains TIR– NBS–lRR (TNl) and WRKY domains, confers immunity on the bacterial pathogen Ralstonia solanacearum by nuclear interaction with the type III bacterial effector PopP2
- 54. AtWRKY52/ RRS1, also interacts with the R protein RPS4 to provide dual resistance towards fungal (Colletotrichum higginsianum) and bacterial pathogens
- 55. AP2/ERF Transcription factor family
- 56. The AP2 domain was first identified as a repeated motif within the Arabidopsis homeotic gene APETALA 2 (AP2) involved in flower development. The ERF domain was first found in tobacco ethylene-responsive element binding proteins (EREBPs) as a conserved DNA-binding motif. AP2/ERF family members are encoded by 145 loci in Arabidopsis and 167 loci in rice. Members of AP2/ERF family are categorized into three subfamilies: the AP2, RAV, and ERF subfamilies. The ERF family is sometimes further classified two major subfamilies, the ERF subfamily and the CBF/ DREB subfamily
- 57. ERF subfamily members are mainly involved in responses to biotic stresses by recognition of the GCC box (5IAGCCGCC3I), which is a DNA sequence involved in ethylene-responsive gene transcription CBF/ DREB subfamily members play crucial roles in abiotic stresses by recognizing a dehydration-responsive element (DRE) with a core motif 5IA/GCCGAC3I. Members of the DREB1/CBF subgroup (DREB1A/CBF3, DREB1B/ CBF1, and DREB1C/CBF2) are cold inducible and are major regulators of cold stress responses, while those of the DREB2 subgroup (DREB2A and DREB2B) play important roles in dehydration and heat stress responses
- 58. Transcription factors without DBD But Interacts with TF with DBD
- 59. AUX / IAA family
- 60. The AUX/IAA family represents a class of proteins interacting with auxin response factors (ARFs). Canonical Aux/IAA proteins share four conserved amino acid sequence motifs known as Domains I, II, III, and IV. Domain I is a repressor domain that contains a conserved leucine repeat motif, similar to the EAR (ethylene-responsive element-binding factor- associated amphiphilic repression) domain Domain I is also required for the recruitment of the transcriptional corepressor TPL C-terminal Domains III and IV are shared with ARF proteins, and are known to promote homo and heterodimerization Domain II confers protein instability, leading to rapid degradation of Aux/IAA through interaction with the f-box protein TIR1
- 63. References
- 64. References Bailey, P.C., Dicks, J., Wang, T.l., Martin, C., 2008. IT3f: a web-based tool for functional analysis of transcription factors in plants. Phytochemistry 69, 2417–2425. Castillon, A., shen, H., Huq, E., 2007. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant sci. 12, 514–521. Deslandes, l., olivier, J., Peeters, N., feng, D.X., Khounlotham, M., Boucher, C., somssich, I., Genin, s., Marco, Y., 2003. Physical inter- action between RRs1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. sci. 100, 8024– 8029. Hahn, A., Bublak, D., schleiff, E., scharf, K.-D., 2011. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell online 23, 741–755. Jakoby, M., Weisshaar, B., Droge-laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., Parcy, f., bZIP Research Group, 2002. bZIP transcription factors in Arabidopsis. Trends Plant. sci. 7, 106–111. Kazan, K., Manners, J.M., 2013. MYC2: the master in action. Mol. Plant 6, 686–703. Li, X., Duan, X., Jiang, H., sun, Y., Tang, Y., Yuan, Z., Guo, J., liang, W., Chen, l., Yin, J., Ma, H., Wang, J., Zhang, D., 2006. Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. Narusaka, M., shirasu, K., Noutoshi, Y., Kubo, Y., shiraishi, T., Iwabuchi, M., Narusaka, Y., 2009. RRs1 and RPs4 provide a dual resistance- gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., shigeoka, s., 2006. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. Ramsay, N.A., Glover, B.J., 2005. MYB–bHlH–WD40 protein com- plex and the evolution of cellular diversity. Trends Plant sci. 10, 63–70.
Editor's Notes
- We will ask some one like this right. What’s the x factor in your success ?
- We will ask some one like this right. What’s the x factor in your success ?
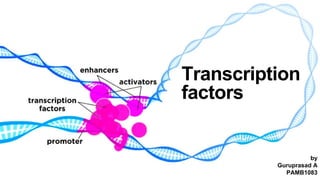
- Here in transcription the factors refers to proteins
- Drosophila 140 mb Cenorhabditis elegans - 100 mb Arabidopsis - 135 mb
- DNA binding domain is the region of TF which goes and binds to the specific DNA recognition sites (Major grove or minor grove), hydrogen bonding with nucleotides, wander val forces etc DNA binding domains classified based on their similarity (similar amino acids sequence chains - which in turn coded by DNA only). Dynamics of chromatin structure, histone variant incorporation, histone eviction.
- GBF – Gbox binding factor
- Myelocytomatosis (similar Homology )
- MYB alone play in cellular morphogenesis, Circadian cycles and organ morphogenesis
- Class B Hsfs are also necessary for suppression of the general Hs response under non-Hs conditions and in the attenuating period.
- Class B Hsfs are also necessary for suppression of the general Hs response under non-Hs conditions and in the attenuating period.