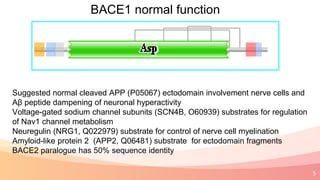
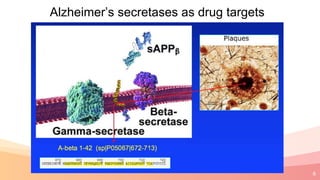
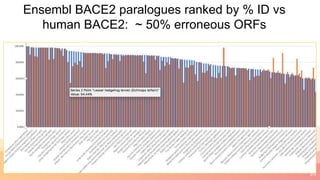
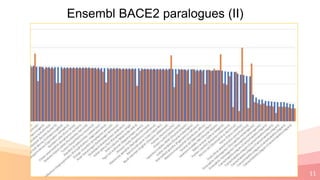
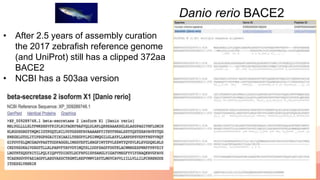
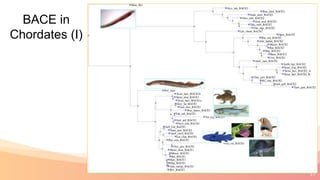
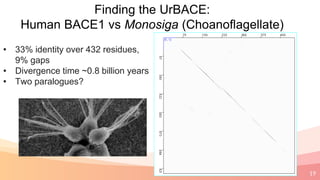
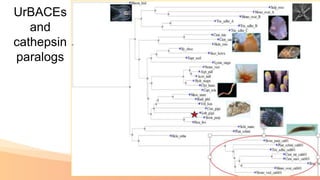
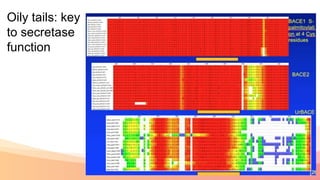
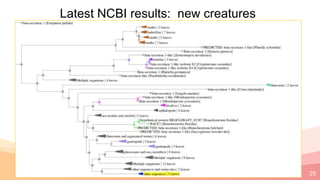
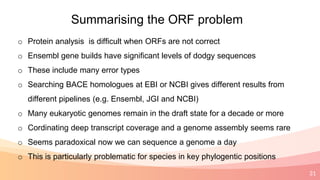
The presentation addresses the challenges in validating bace1 and bace2 as drug targets for Alzheimer's and diabetes, highlighting issues with incorrect protein sequences and unfinished representative genomes. It discusses the phylogenetic analysis of these proteases, the evolution of bace enzymes, and the prevalence of errors in genomic data. The document also proposes potential solutions and raises questions for future research in this field.