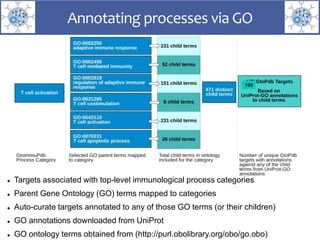
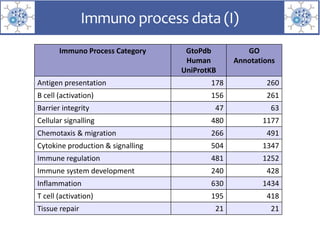
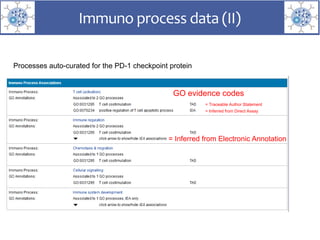
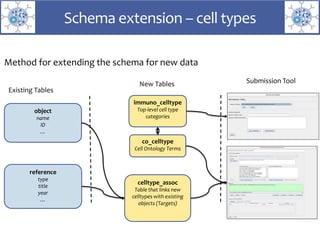
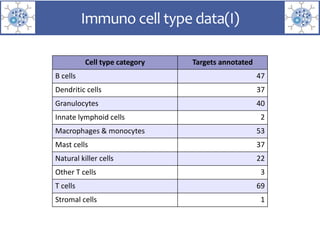
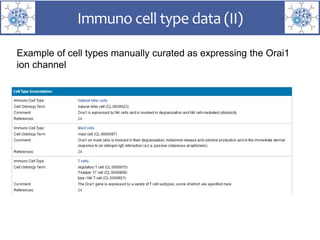
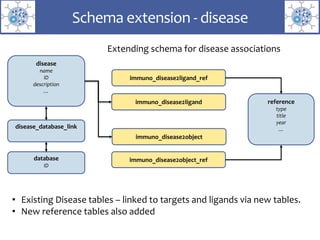
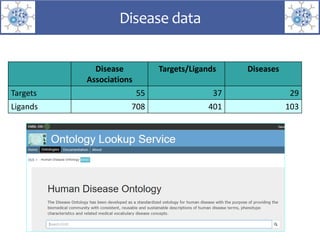
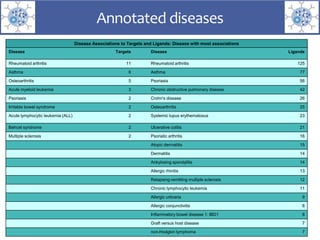
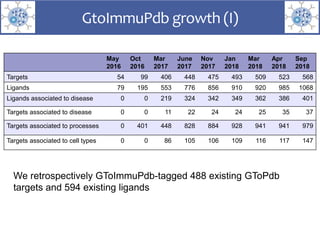
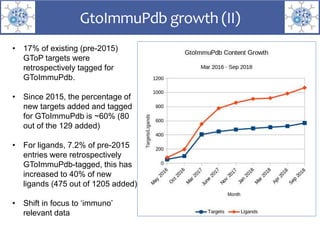
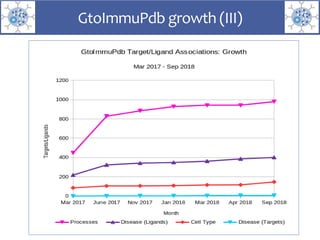
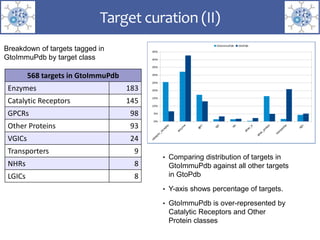
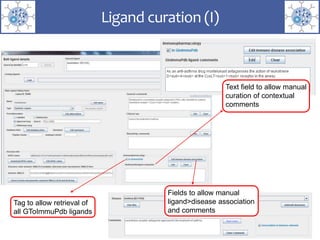
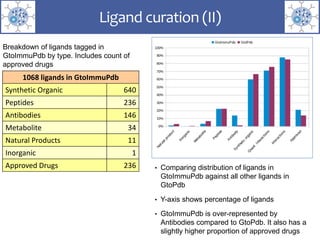
The presentation outlines the development of the GtoImmupDB, an extension of the existing GtoPdb database focusing on immunopharmacology. New data types, including processes, cell types, and diseases, have been integrated, along with literature sourcing and curation methods highlighted for capturing relevant references. Key statistics on targeted and ligand data are included, underscoring the challenges of quality assessment in immunopharmacology and emphasizing the need for continued collaborative efforts and expert feedback.






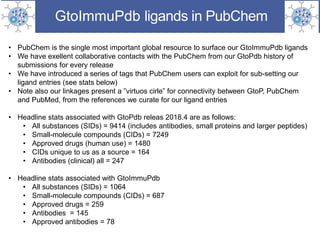
![PubChem antibodies
• Substance side query: "IUPHAR/BPS Guide to PHARMACOLOGY"[SourceName]” AND
”immunopharmacology” AND “approved” AND “antibody”](https://image.slidesharecdn.com/gtoimmupdboct2018-181008092854/85/Guide-to-Immunopharmacology-update-14-320.jpg)
![PubChem small-molecules
• Query "IUPHAR/BPS Guide to PHARMACOLOGY"[SourceName]” as CIDs from
“immunopharmacology, select for unique to us and sorted by date](https://image.slidesharecdn.com/gtoimmupdboct2018-181008092854/85/Guide-to-Immunopharmacology-update-15-320.jpg)