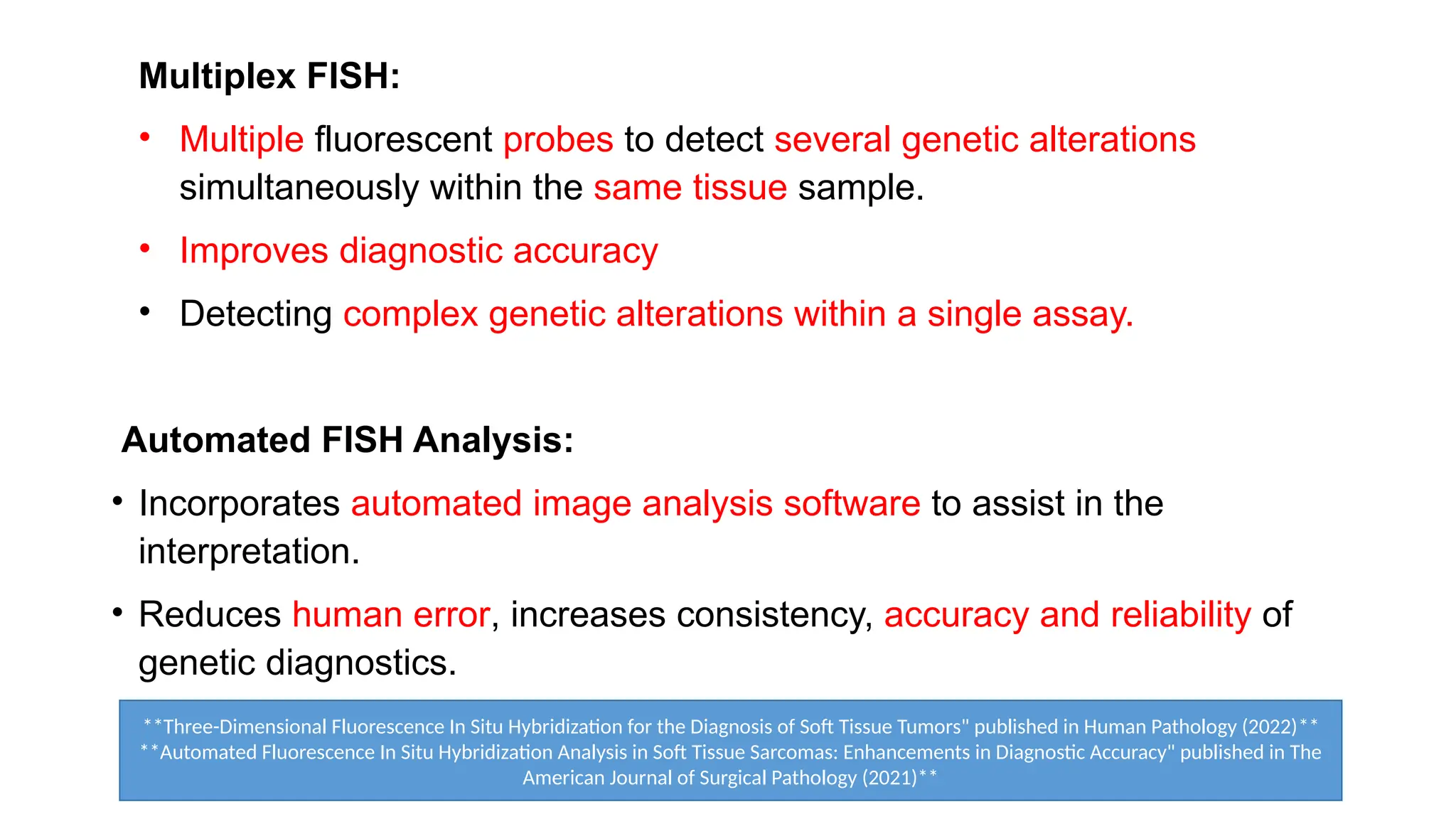
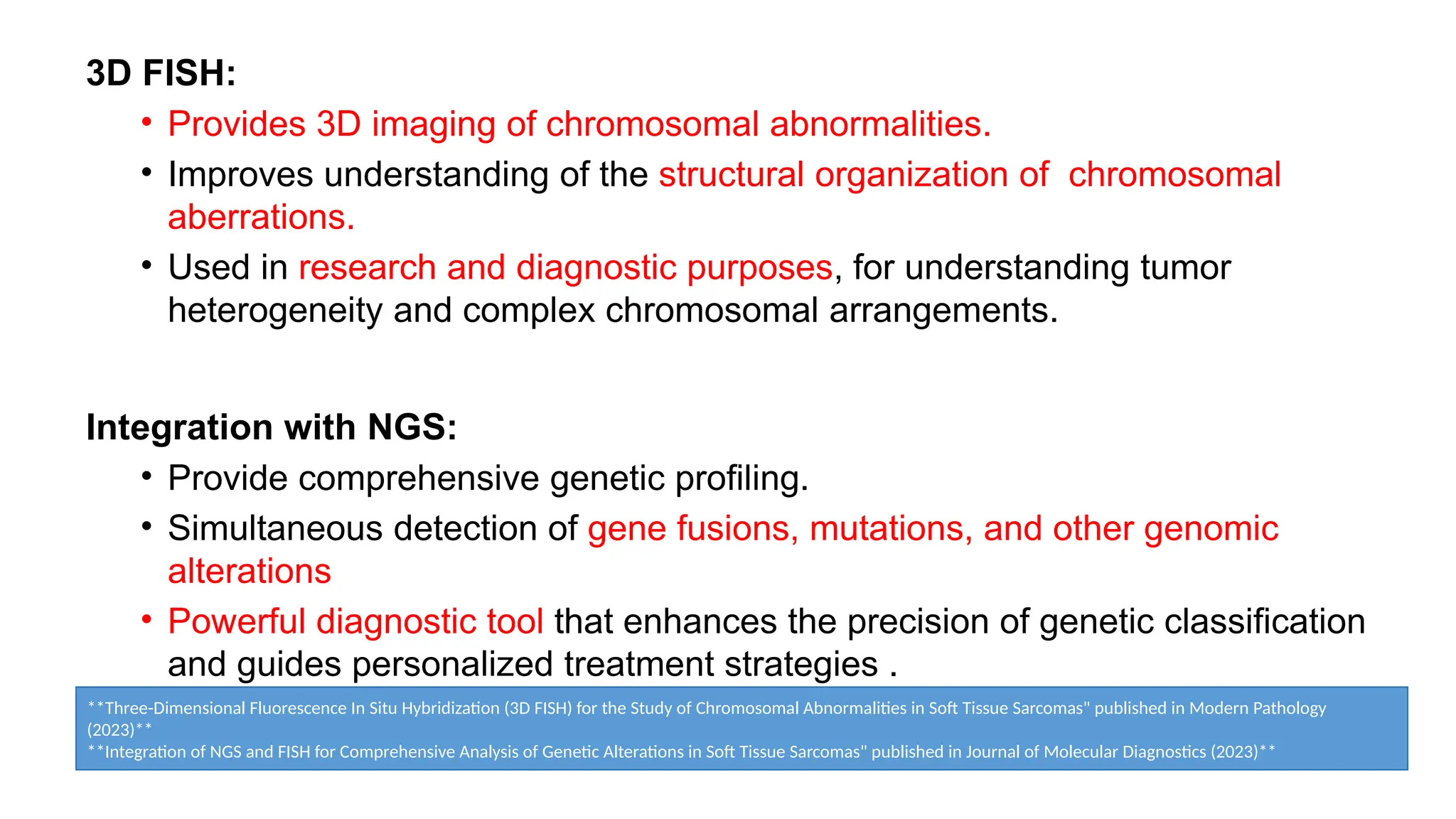
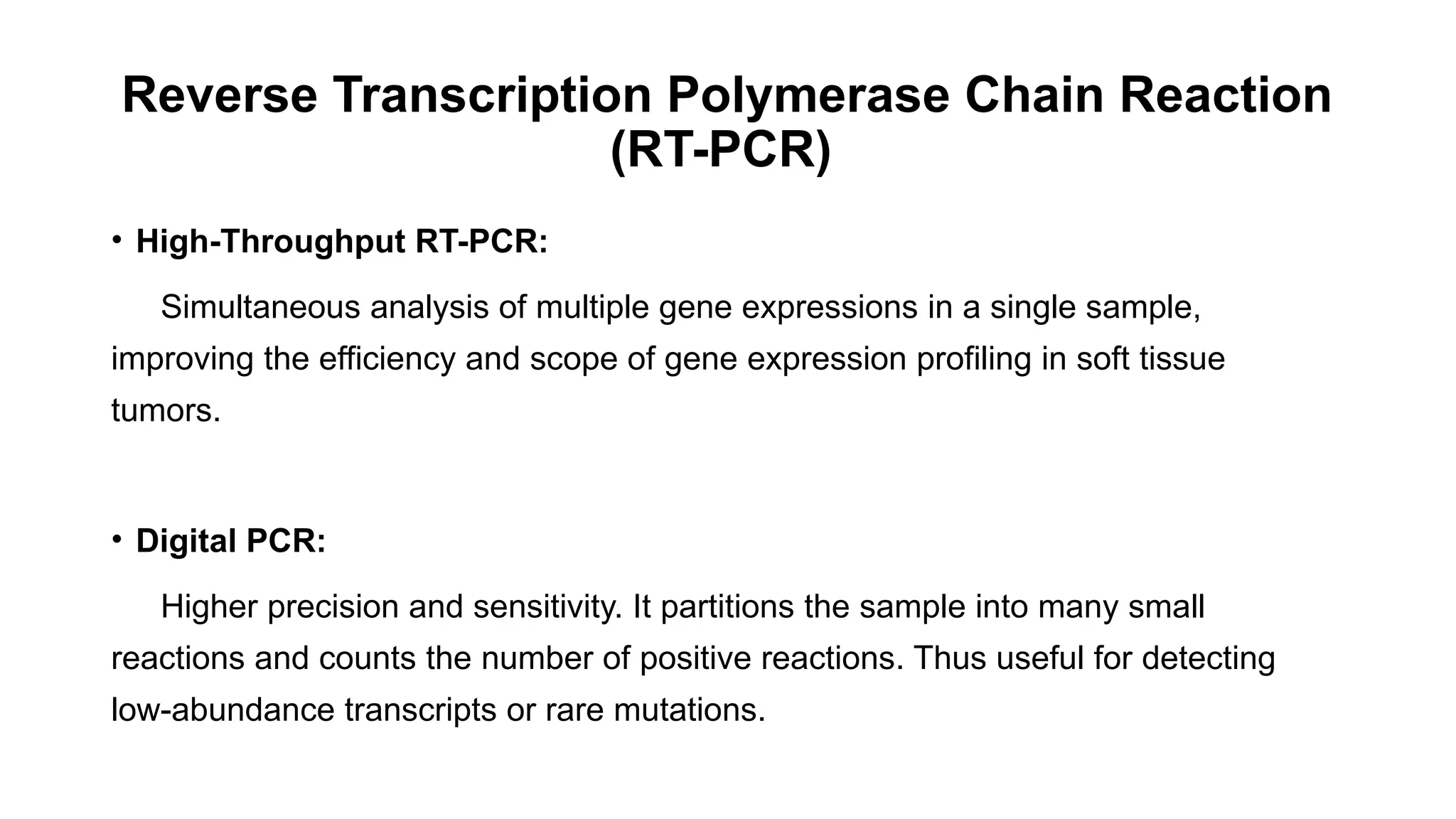
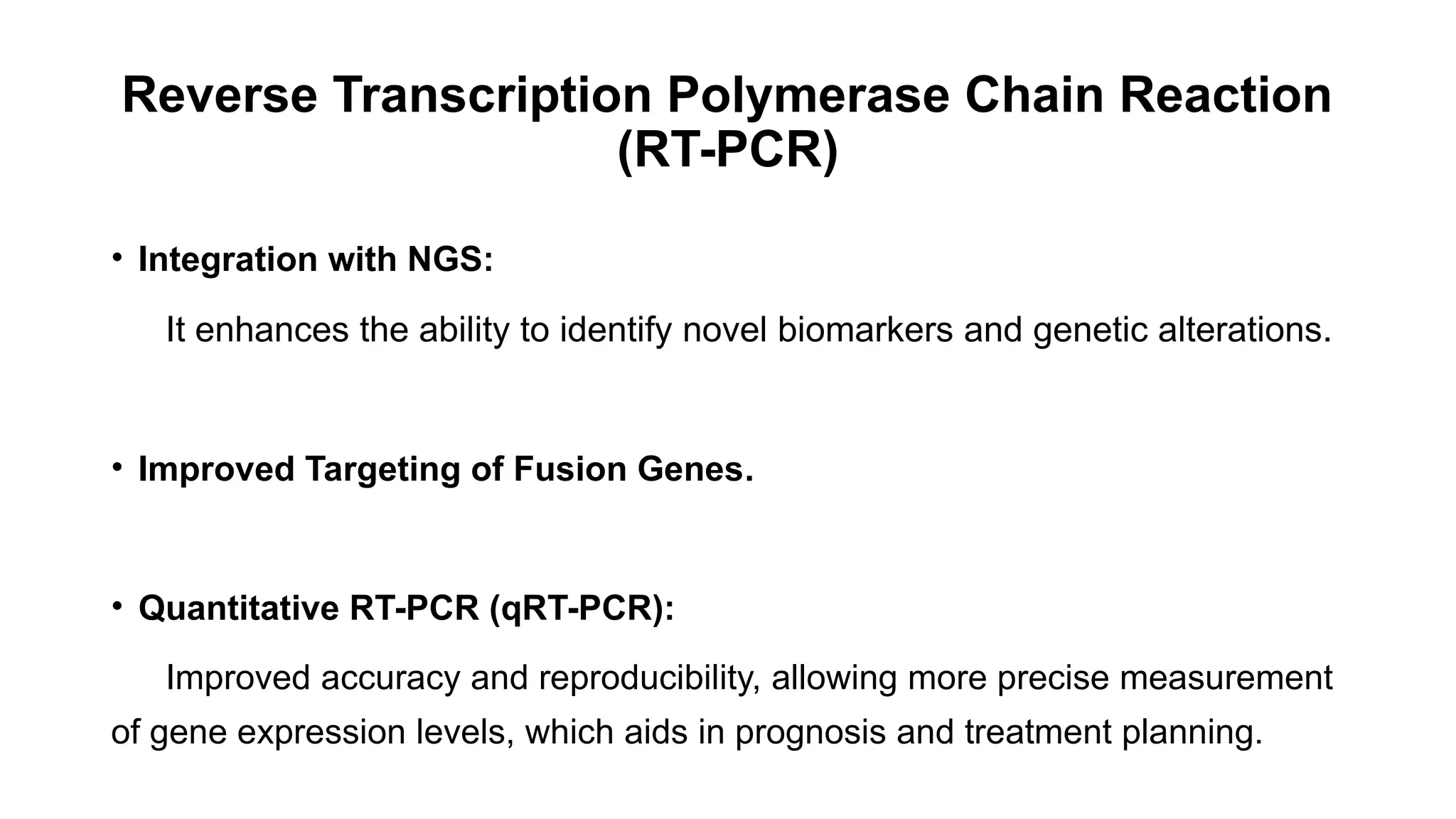
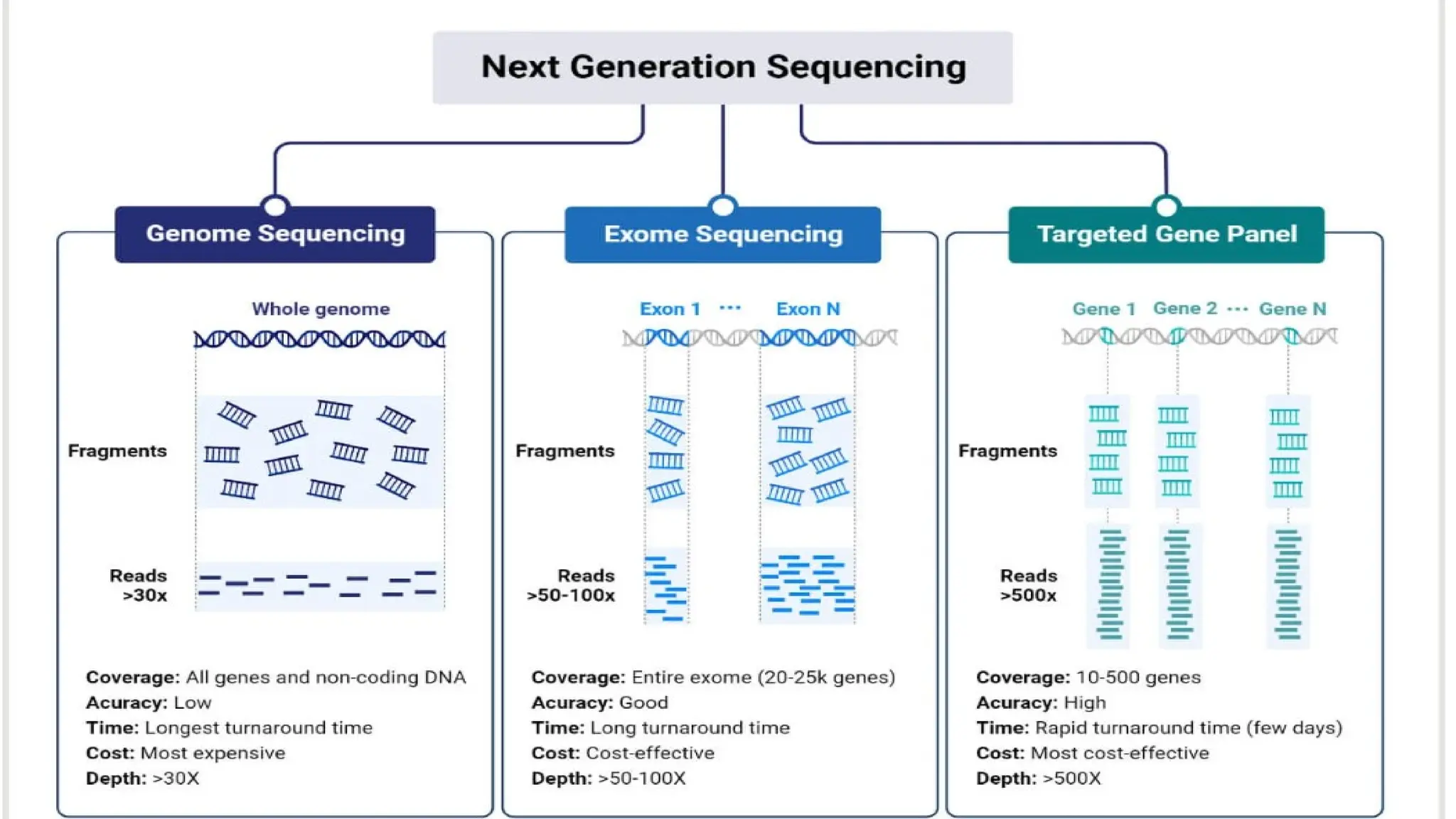
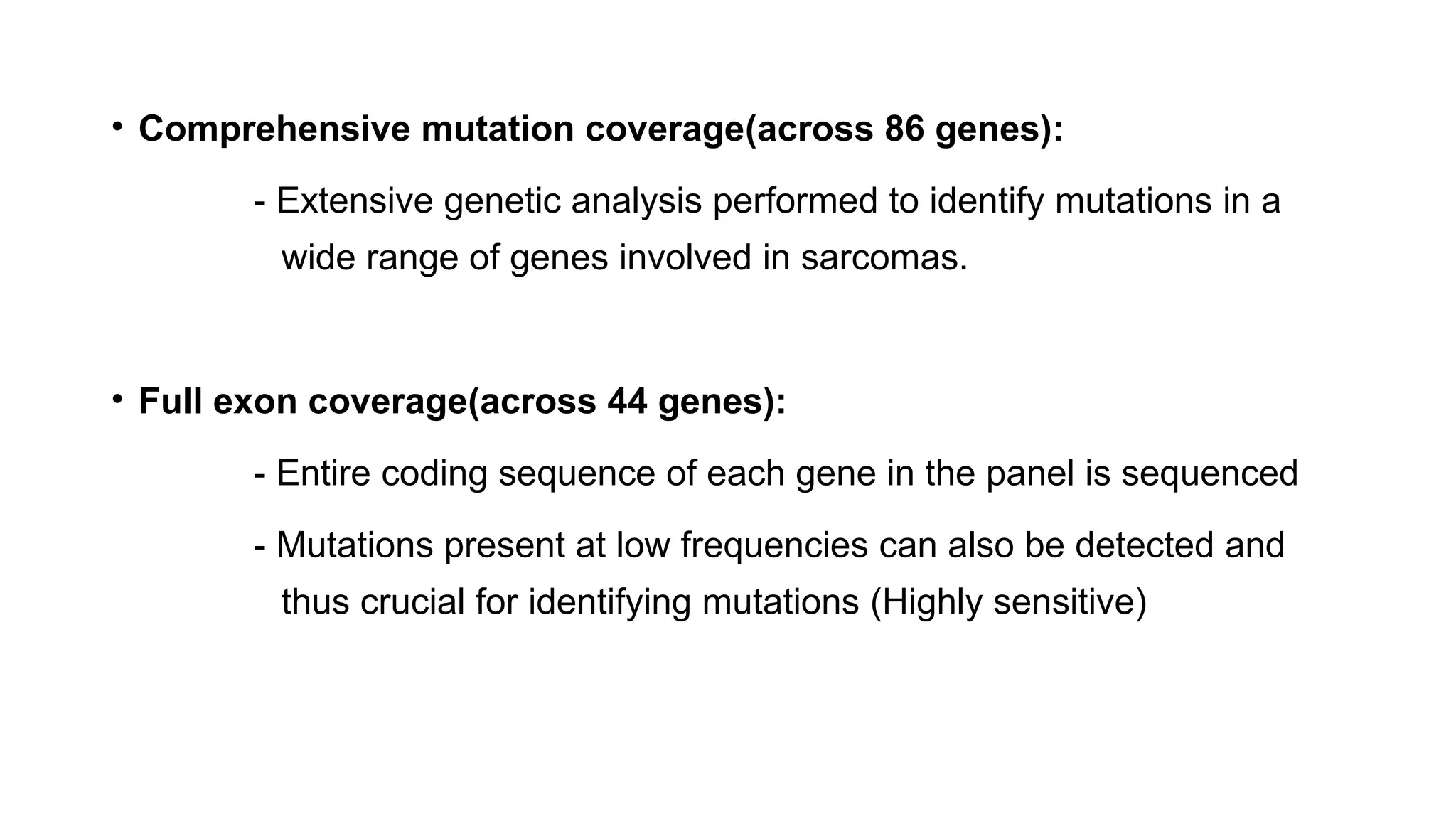
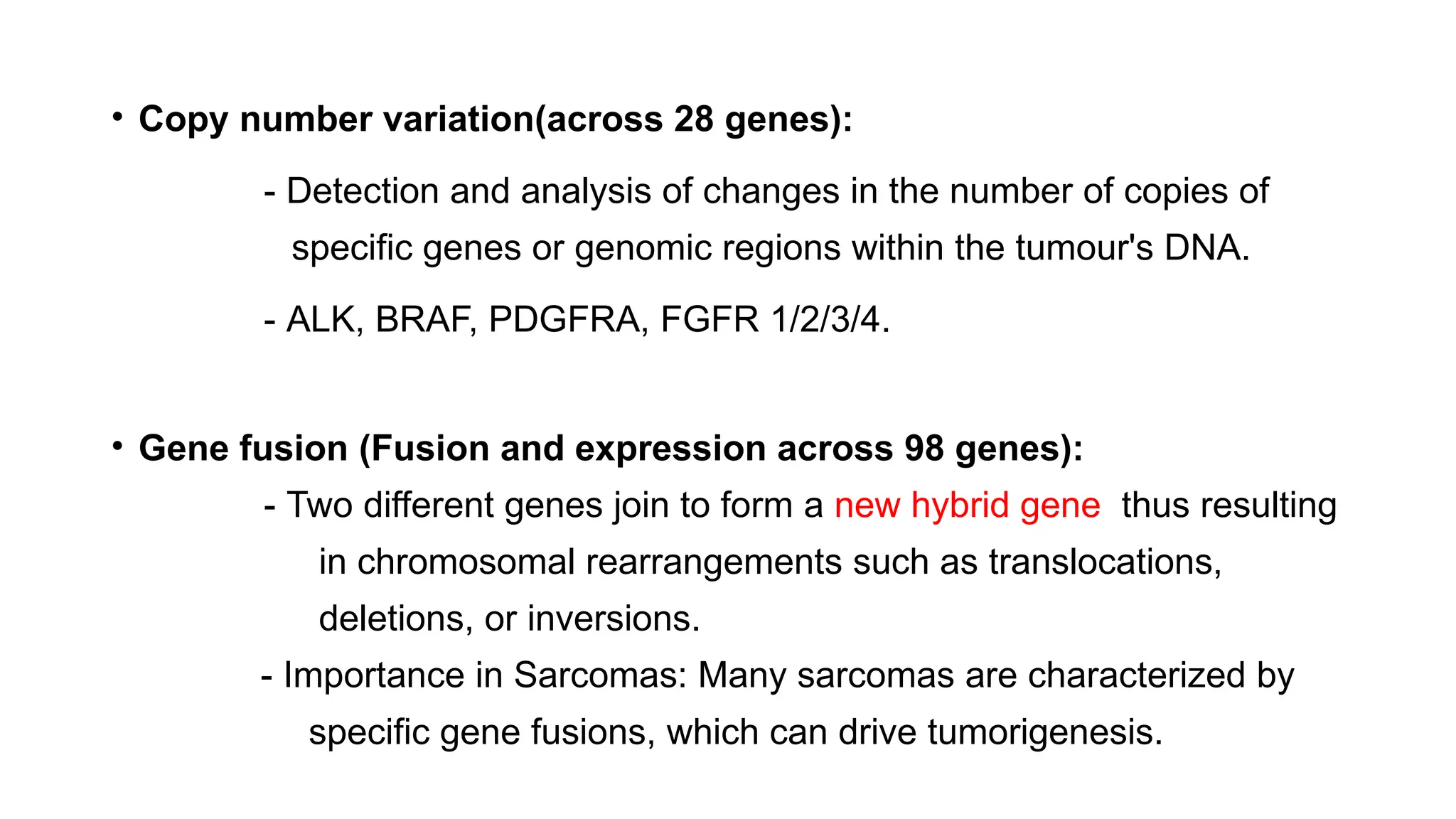
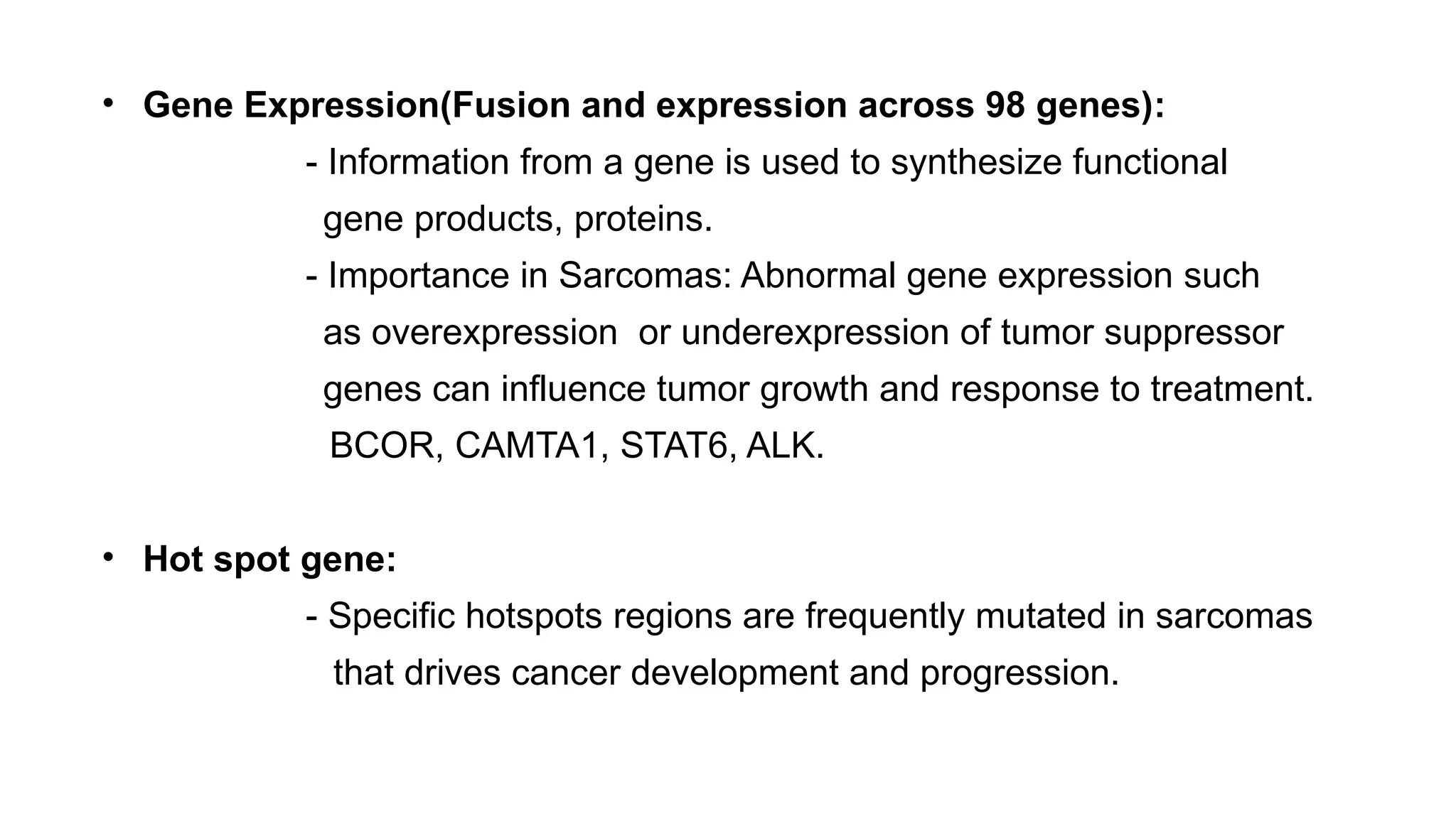
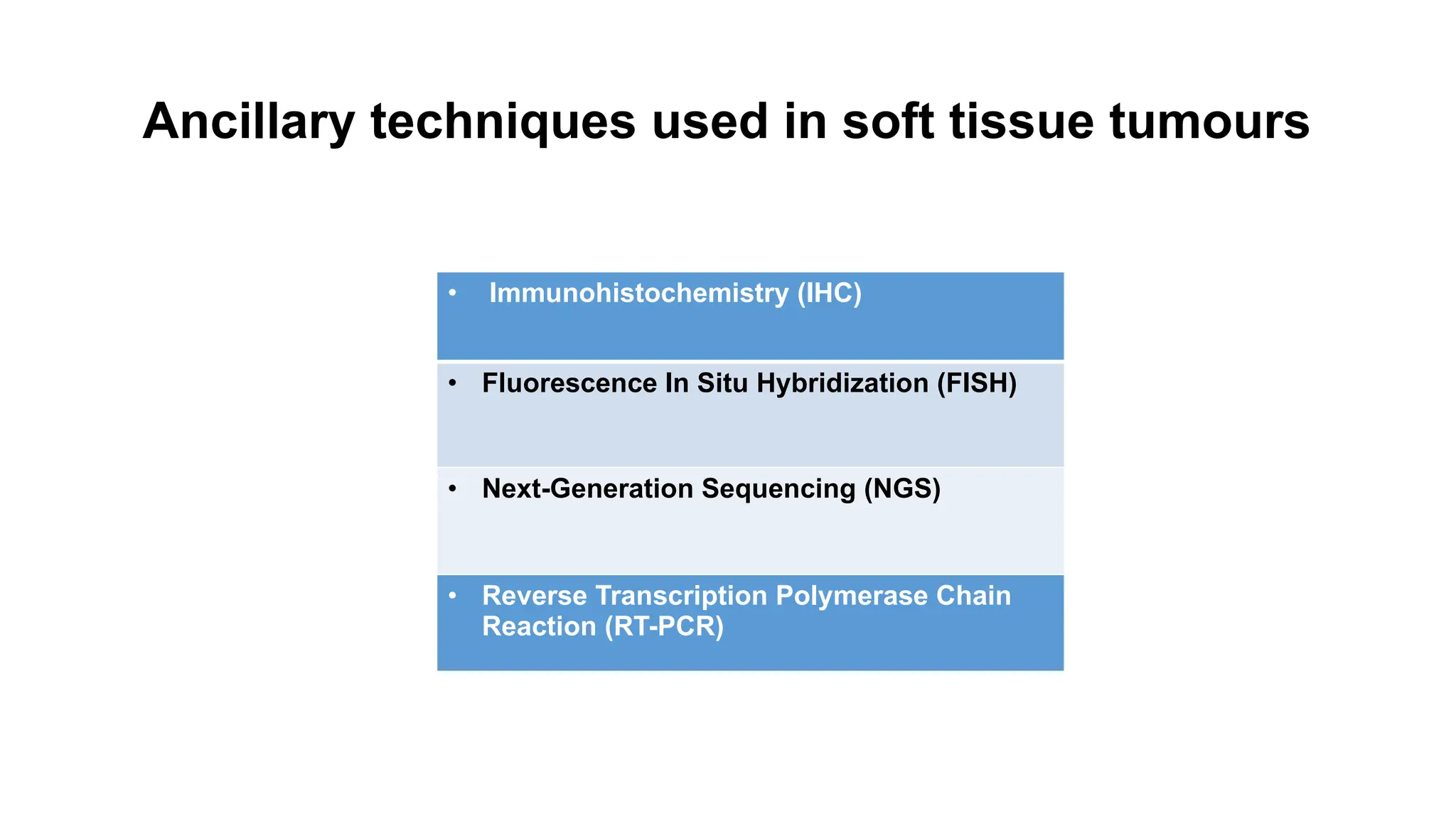
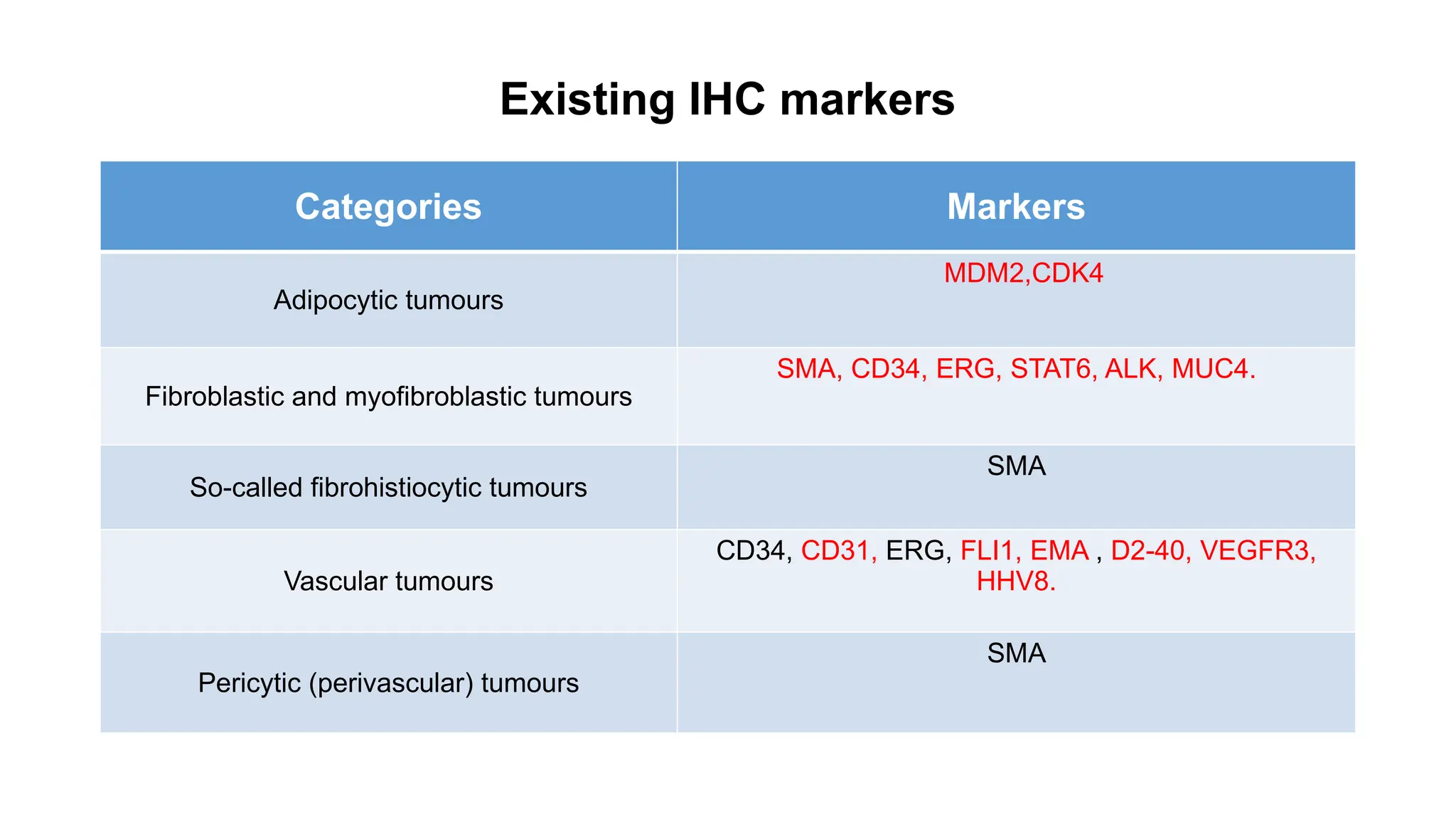
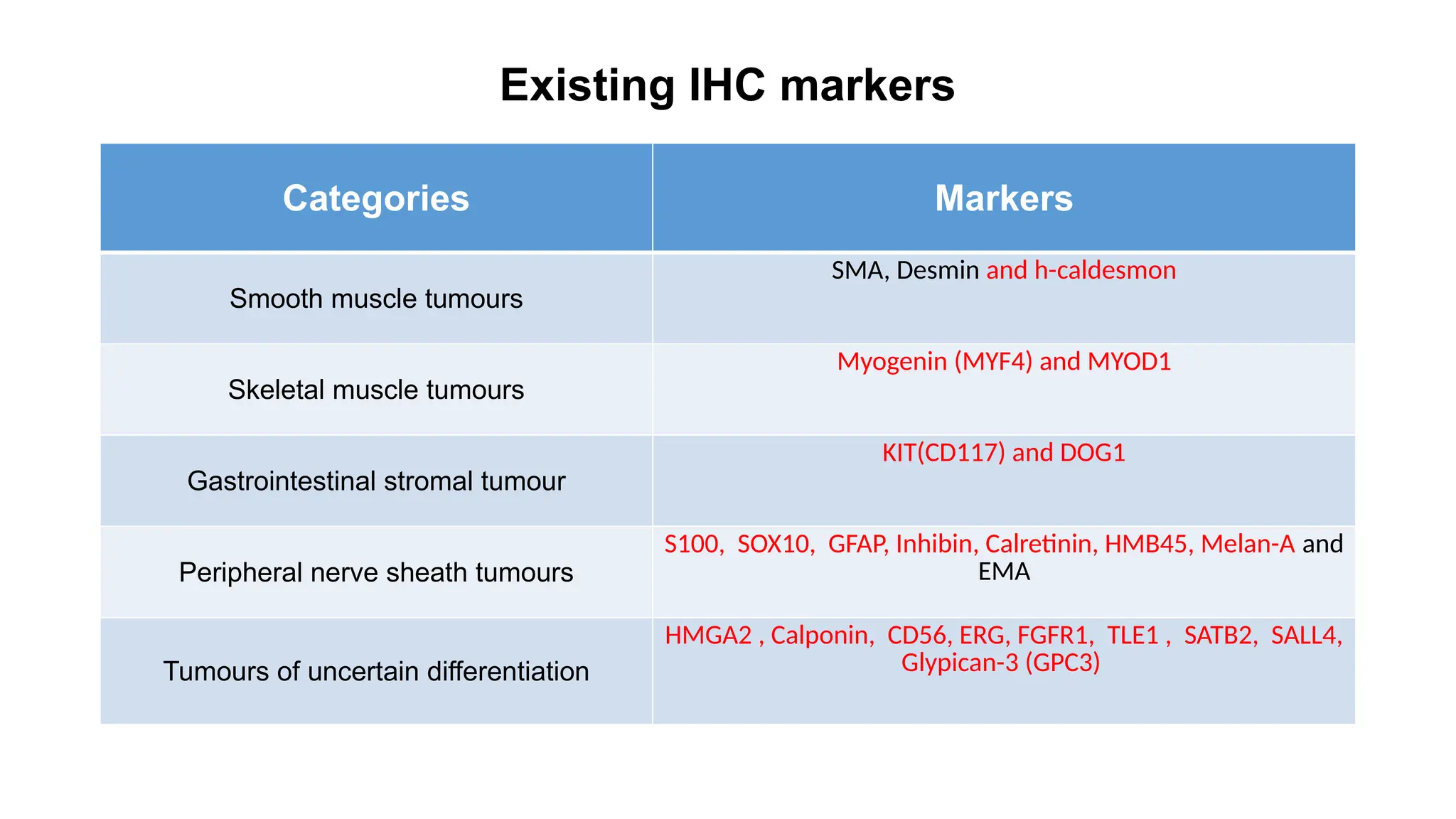
The document discusses recent advancements in ancillary techniques for diagnosing soft tissue sarcomas, highlighting a range of immunohistochemical (IHC) and molecular techniques including RT-PCR, FISH, and next-generation sequencing (NGS). It emphasizes the importance of combining morphologic analyses with molecular pathology to enhance diagnostic accuracy and guide personalized treatment strategies. Additionally, the role of artificial intelligence in improving diagnostic precision and treatment decision-making for sarcomas is detailed.







![CAMTA1: Calmodulin-binding transcription activator 1
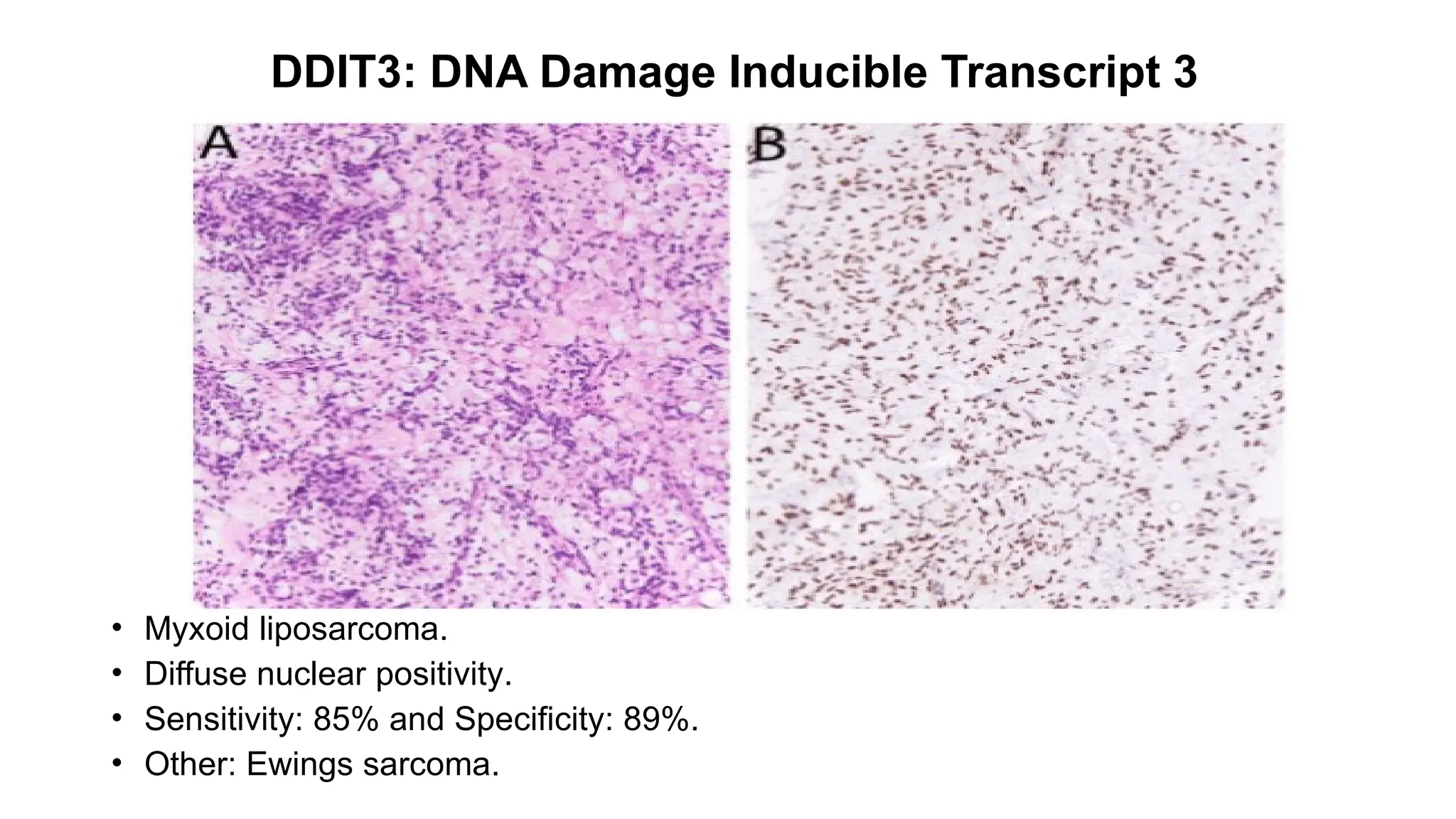
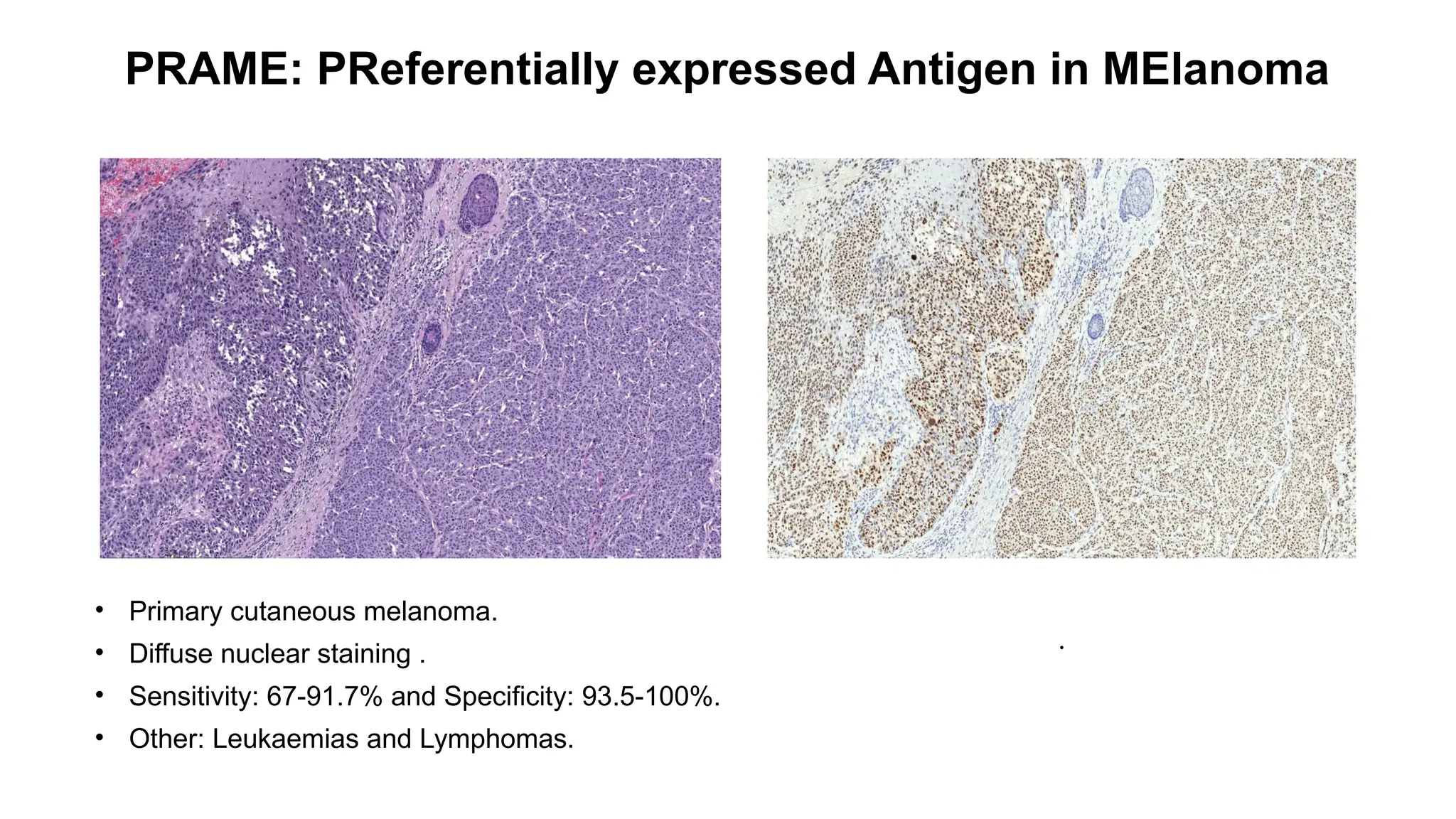
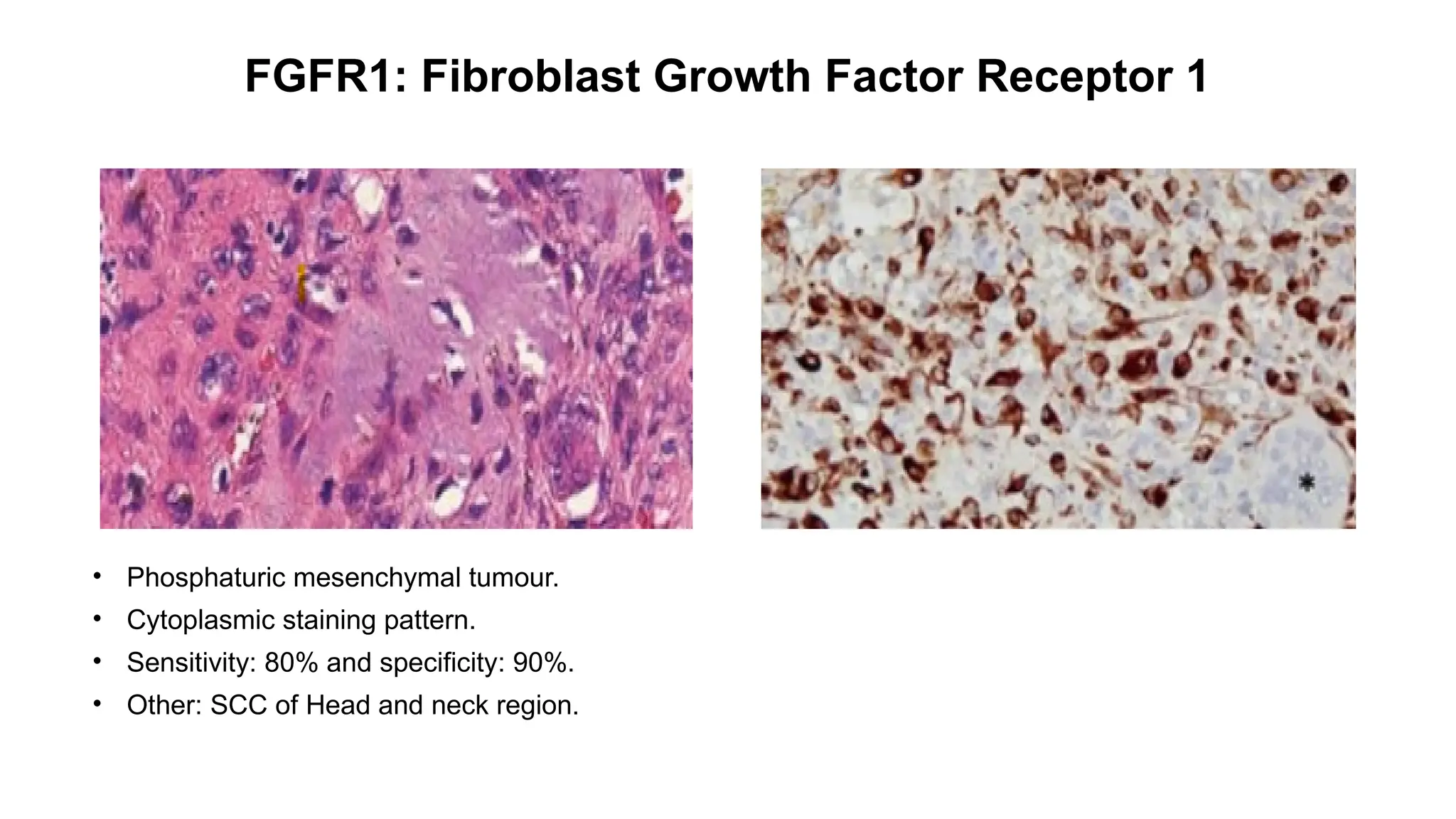
• Epithelioid hemangioendothelioma [WWTR1::CAMTA1 (85–90%) ]
• Strong nuclear.
• Sensitivity: 91.2% and Specificity: 100%.
• Other: Gliomas](https://image.slidesharecdn.com/123456789-1-240920021608-1463d67e/75/Recent-advances-in-soft-tissue-sarcoma-and-its-applications-14-2048.jpg)




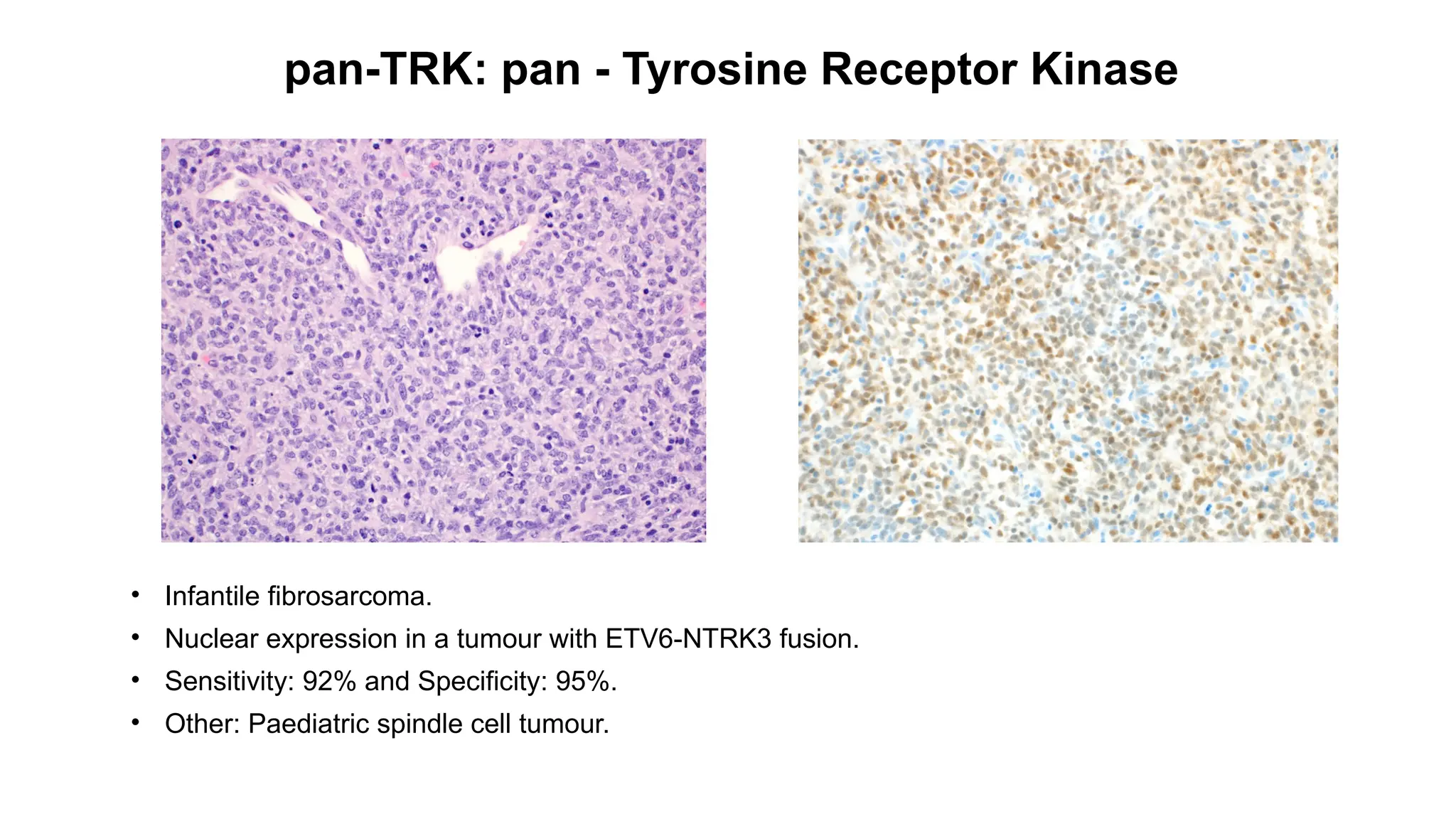
![DUX4
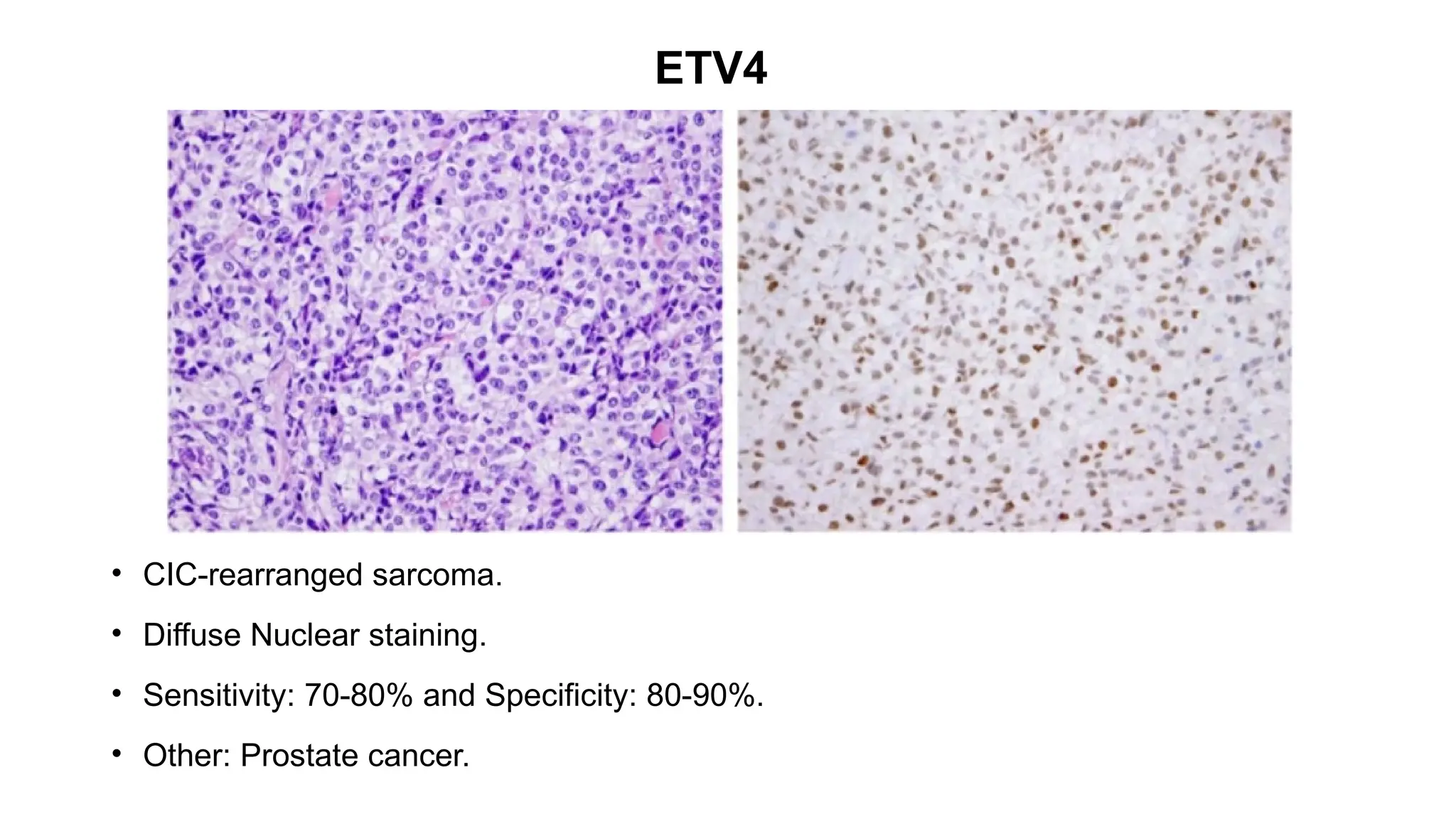
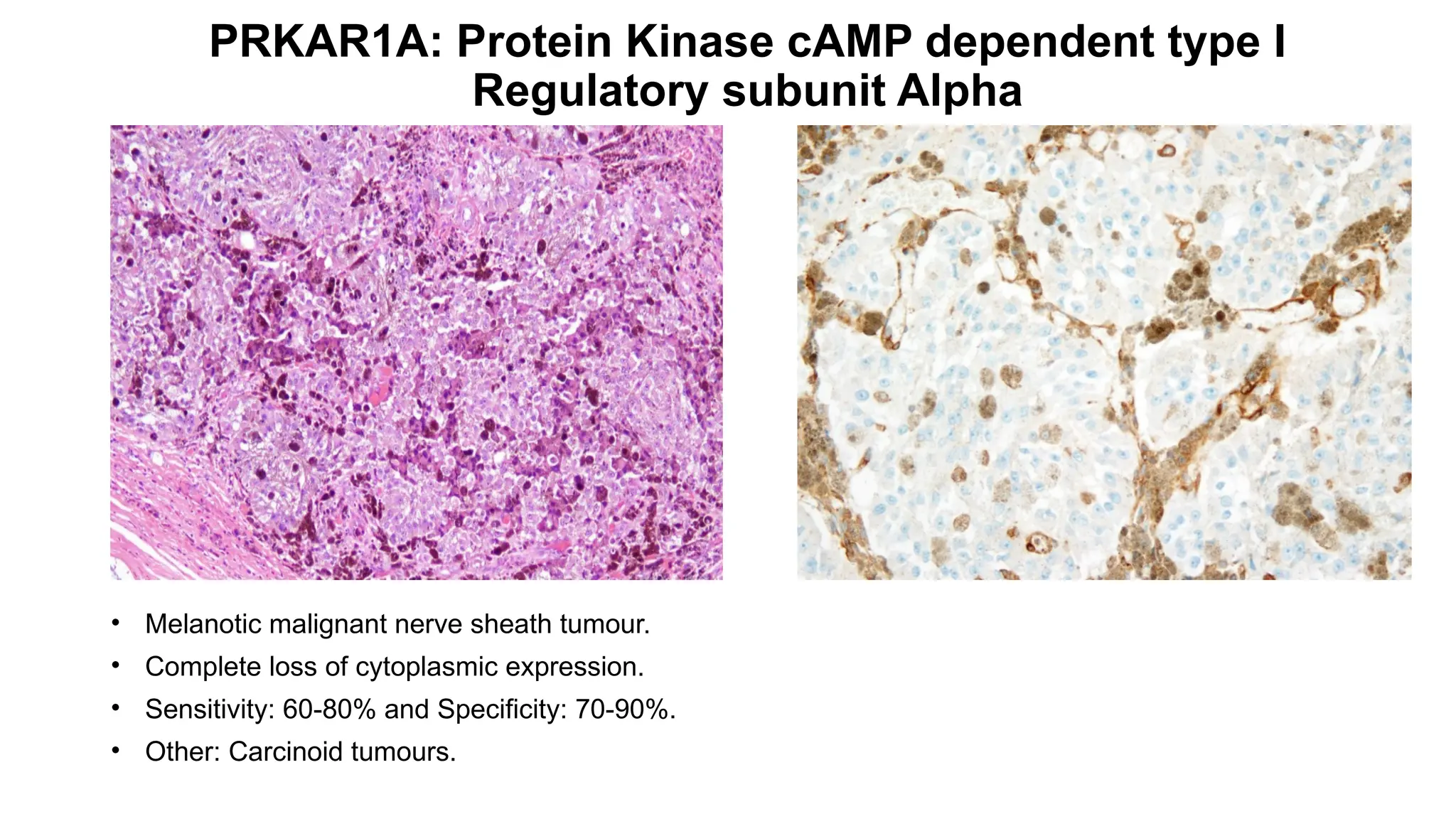
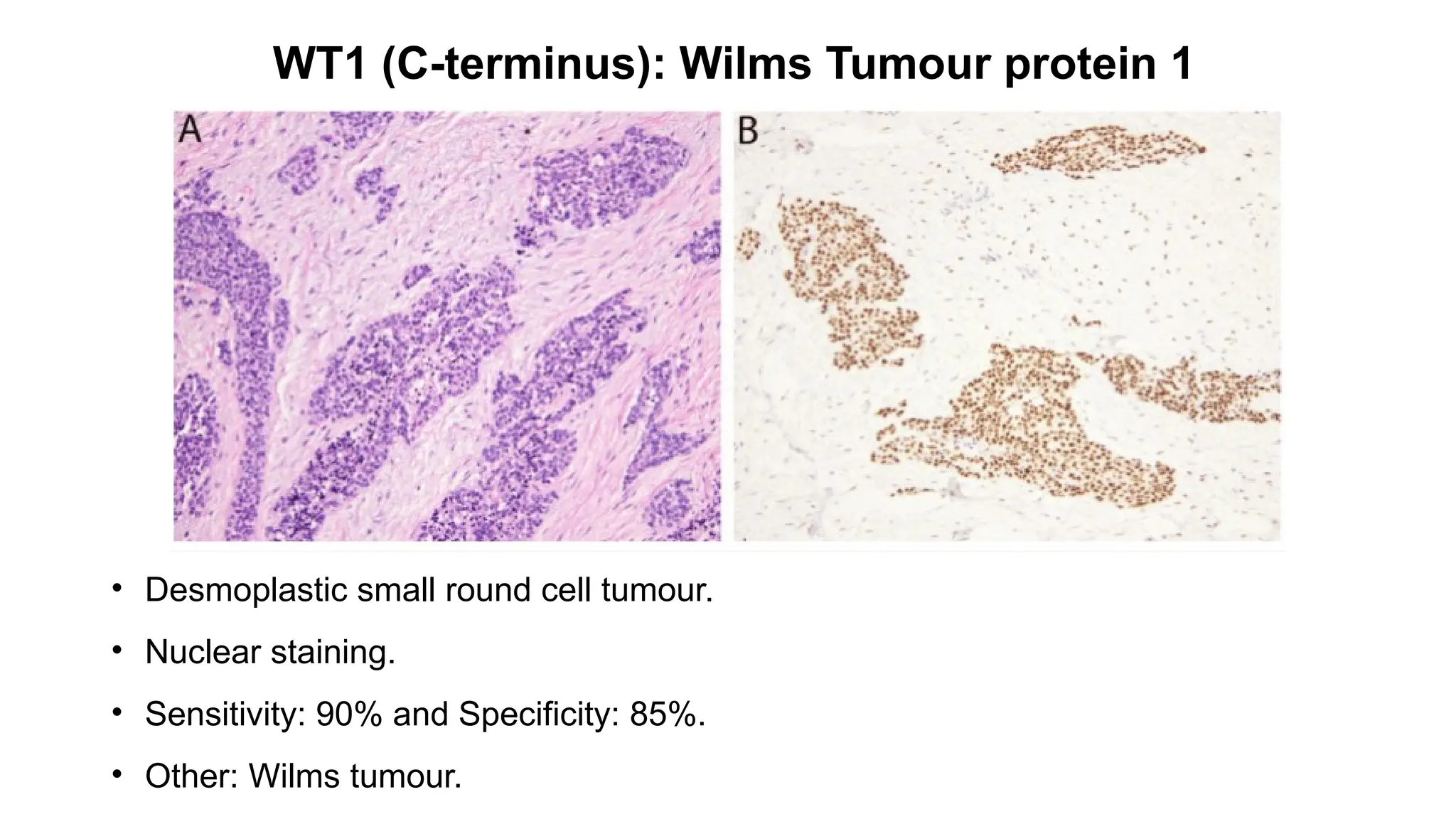
• CIC-rearranged sarcoma[CIC::DUX4 (95%)].
• Nuclear staining.
• Sensitivity: 90% and Specificity: 85% .
• Other: Ewing sarcoma, Desmoplastic small round cell tumour.](https://image.slidesharecdn.com/123456789-1-240920021608-1463d67e/75/Recent-advances-in-soft-tissue-sarcoma-and-its-applications-26-2048.jpg)