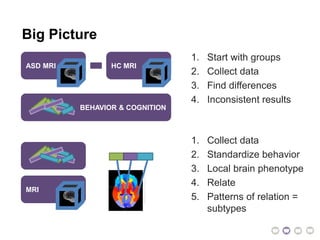
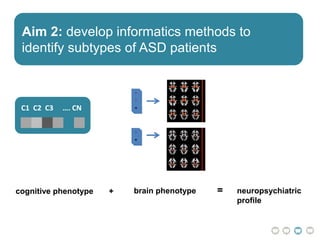
The document outlines a hypothesis regarding the reattribution of well-known semantic verse terms to express mutual sentiments within a feature group, presented by Vanessa Sochat. It also details a plan for a computational study aiming to develop informatics methods for identifying subtypes of autism spectrum disorder (ASD) from imaging and behavioral data to enhance early diagnosis and tailored treatment. Key contributions include biological markers for ASD and decision support tools for treatment based on neural profiles.