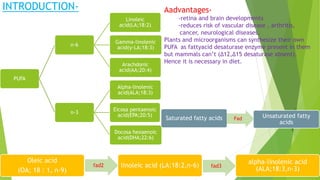
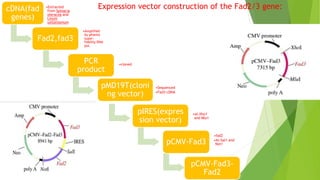
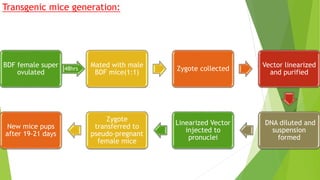
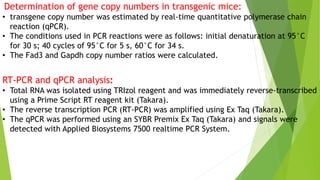
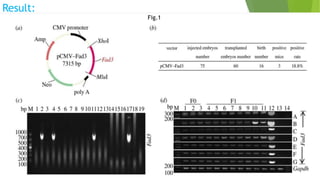
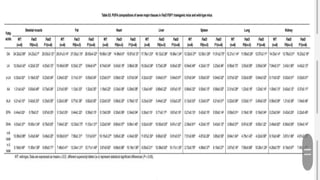
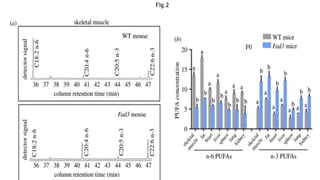
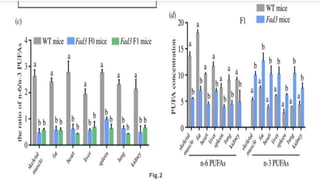
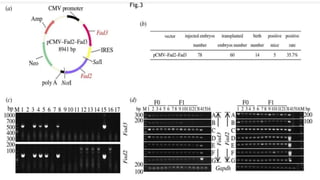
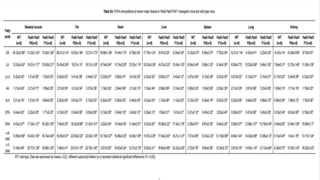
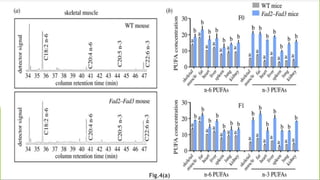
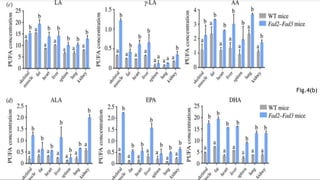
This research paper describes the generation of transgenic mice expressing plant-derived fatty acid desaturase (Fad) genes. The Fad2 and Fad3 genes from spinach and flax were introduced separately and together into mice via pronuclear microinjection. Mice expressing Fad3 alone or both Fad2 and Fad3 were able to endogenously synthesize the polyunsaturated fatty acids linoleic acid and α-linolenic acid, demonstrating that plant Fad genes can be functionally expressed in mammals. This establishes a method for in vivo production of these essential fatty acids without requiring them in the diet.