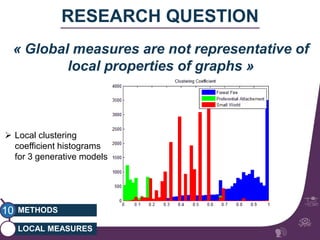
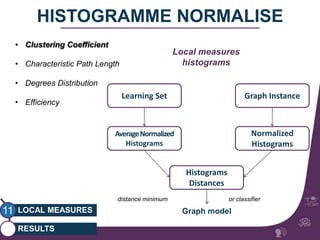
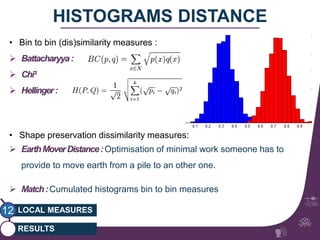
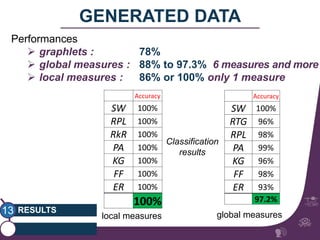
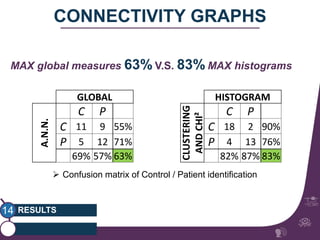
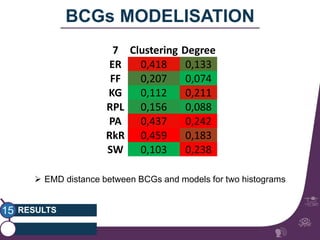
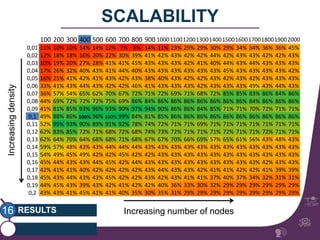
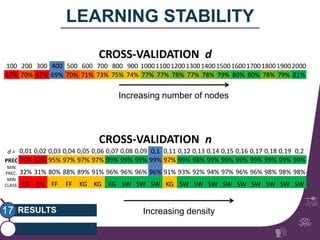
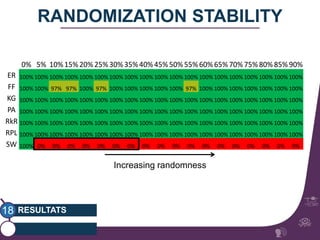
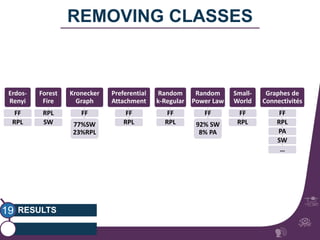
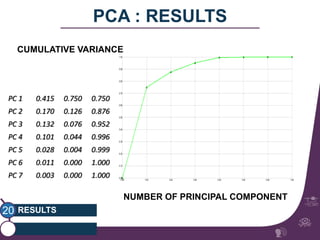
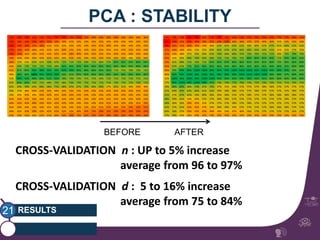
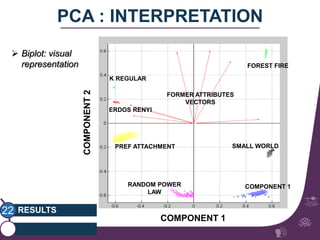
This document summarizes a study on classifying brain connectivity graphs (BCGs) using graph theory measures. It introduces graph theory concepts and previous work using global and local measures. The study finds that using histograms of local clustering coefficients and chi-squared distance achieves 83% accuracy classifying BCGs, outperforming global measures. It also examines the stability, scalability and effects of randomization on the classification approach.