Petteri Teikari provides a shallow literature analysis of asthma diagnostics and management to aid in developing digital solutions. Asthma is heterogeneous with multiple endotypes requiring personalized treatment. Over and underdiagnosis are common due to a lack of objective lung function testing to demonstrate variable airflow limitation supporting an asthma diagnosis. Effort-free lung function measures are desired. Asthma is an umbrella term for various subtypes that are managed through preventer inhalers, reliever inhalers, and action plans tailored to the individual.











![Asthma Basics #2
NICE guideline [NG80] UK (March 2021): “Asthma: diagnosis, monitoring
and chronic asthma management”](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-13-2048.jpg)












![Obesity-related asthma #2
View of the hormonal regulation of airway responsivenes state, adiponectin plays an
anti-inflammatory role, while leptin n (b) In obesity/insulin resistant condition, there is a
reduction in a levels, which in turn plays a pro-inflammatory role in the lungs airways.
Ach: acetylcholine; LR: leptin resistance. - Leiria et al. (2015): “Obesity and asthma:
beyond T(H)2 inflammation.”
Yu et al. (2023): “The relationship between the use
of GLP-1 receptor agonists and the incidence of
respiratory illness: a meta-analysis of randomized
controlled trials”
GLP-1 RAs may reduce asthma through
weight loss and improved insulin
sensitivity [Cardet et al. 2016, Rodrigues et al. 2017,
Rodrigues et al. 2020]
. In addition, in a study that
included 16,690 patients, patients with
GLP-1RA experienced less worsening of
chronic lower respiratory disease
compared to DPP-4I users [Albogami et al. 2021]
The treatment — semaglutide
(Ozempic®, Novo Nordisk) ... “Our trial
represents the first prospective study of
this class of medications in individuals
with asthma,”](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-32-2048.jpg)











![Bronchodilator reversibility Spirometry
Ahmed et al. (2023): “Bronchodilator reversibility testing in morbidly obese non-
smokers: fluticasone/salmeterol efficacy versus salbutamol bronchodilator”
A positive response in reversibility testing is widely used to diagnose patients
with airway limitations. However, despite its simple procedure, it doesn’t accurately
reflect the exact airway irreversibility. This study aimed to investigate the efficacy of a
bronchodilation reversibility test using salbutamol and fluticasone/salmeterol
combination in obese non-smoker subjects.
Spirometry is pivotal in assessing bronchodilator reversibility. It is considered the
standard gold standard for diagnosing diseases with obstructive airways in general
practice. Reversibility testing measures the airflow expiration response after
inhaling a bronchodilator. A positive test is defined as an increase in FEV1 of more
than 12% from baseline or a 200-ml increase, according to the American Thoracic
Society (ATS). In comparison, the European Respiratory Society (ERS) recommended a
change of more than 9% of the predicted FEV1 as a hallmark for asthma confirmation
[1]. However, its results have limited reproducibility and accuracy. Since it depends
on several factors, such as the patient’s maximal effort and the bronchodilator
used [3].
Fluticasone/salmeterol combination increases FEV1, FEV1% of predicted, and FEV1/FVC
ratio than the conventional test using salbutamol inhaler, and it can be a potential
candidate for assessment of airway obstruction using reversibility test, especially
among the obese population.
Visser et al. (2015): “Reversibility of pulmonary function after inhaling
salbutamol in different doses and body postures in asthmatic children”](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-48-2048.jpg)

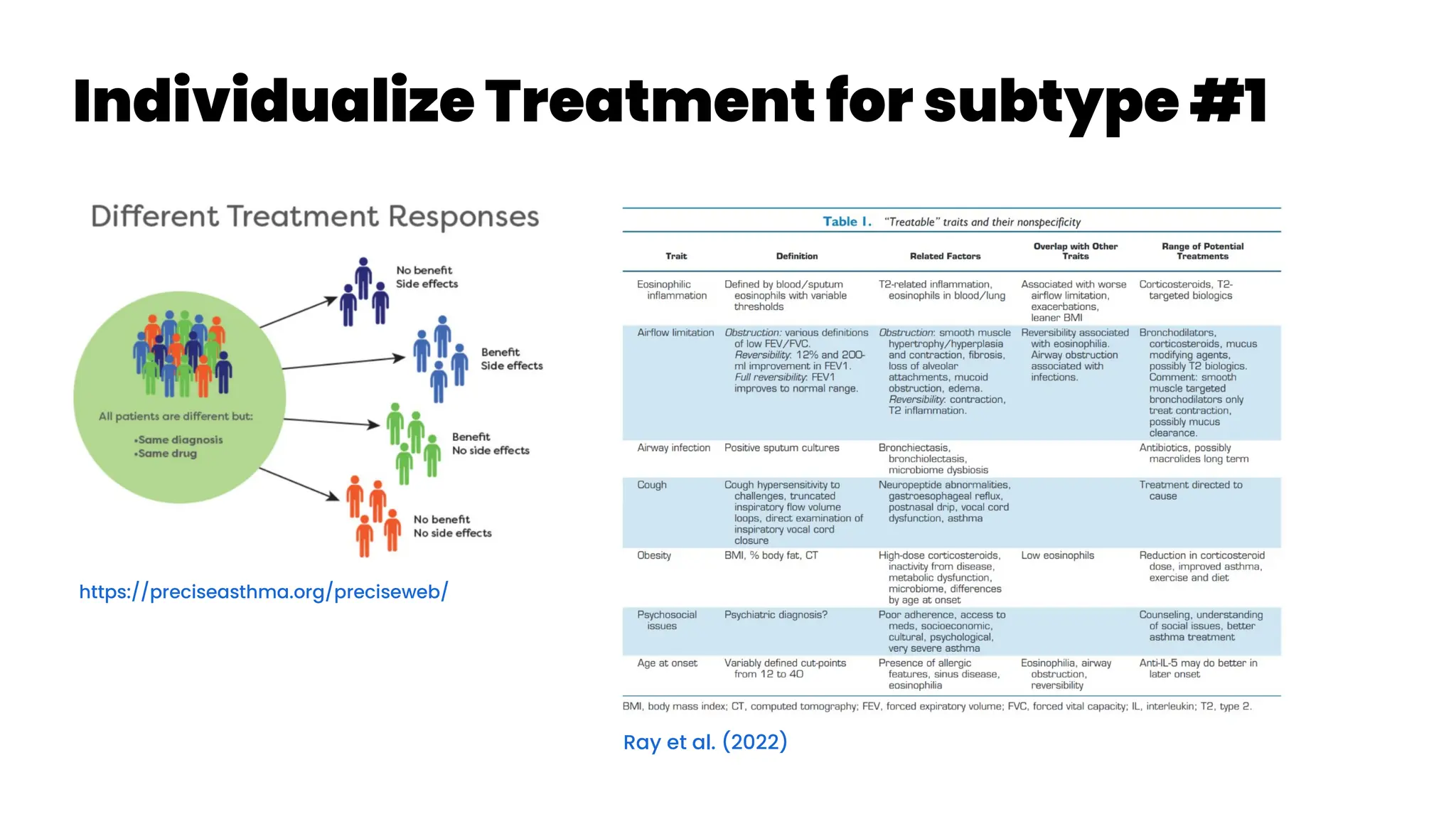







![Respiratory Biomarkers #5
Leung and Sin (2013): “Biomarkers in airway
diseases”
The inherent limitations of spirometry and clinical
history have prompted clinicians and scientists to
search for surrogate markers of airway diseases.
Although few biomarkers have been widely
accepted into the clinical armamentarium, the
authors explore three sources of biomarkers that
have shown promise as indicators of disease
severity and treatment response. In asthma, exhaled
nitric oxide measurements can predict steroid
responsiveness and sputum eosinophil counts have
been used to titrate anti-inflammatory therapies. In
chronic obstructive pulmonary disease, inflammatory
plasma biomarkers, such as fibrinogen, club cell
secretory protein-16 and surfactant protein D, can
denote greater severity and predict the risk of
exacerbations. While the multitude of disease
phenotypes in respiratory medicine make biomarker
development especially challenging, these three may
soon play key roles in the diagnosis and
management of airway diseases.
Papi et al. (2021): “Rate of Decline of FEV1 as a Biomarker of
Survival?”
COPD has been defined by the excess decline in lung
function induced by tobacco smoking, with FEV1
considered the gold standard biomarker of COPD
development and progression
Dobler (2019): “Biomarkers in respiratory diseases”
The December issue of Breathe focuses on biomarkers in
respiratory diseases [Turner et al. 2019; Oliver et al. 2019;
Creamer et al. 2019; Hamerlijnck et al. 2019]. Biomarkers are
measurable indicators of the presence, severity or type of
a disease. They can help us understand the cause,
phenotype, progression or regression, prognosis, or outcome
of treatment of a disease. Biomarkers hold the promise of
personalised medicine, which aims to tailor treatments to
individual patients based on their biomarker profile and, by
doing so, reduce the harms from ineffective treatments
and increase the benefits from effective treatments.](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-60-2048.jpg)







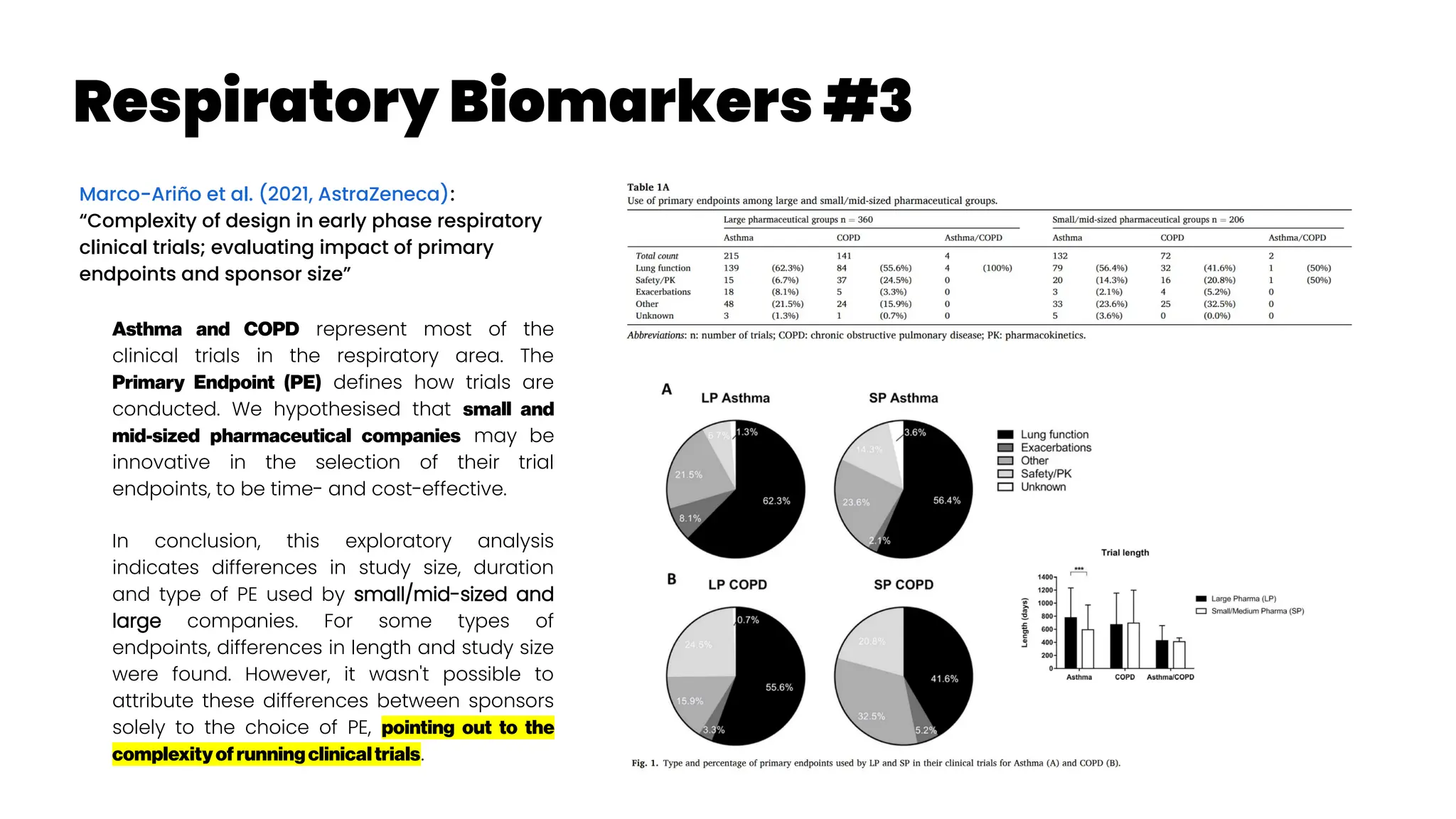
![EHR for asthma management #1
Alharbi et al. (2021): “Predictive models for
personalized asthma attacks based on
patient’s biosignals and environmental
factors: a systematic review”
Many researchers developed asthma attacks
prediction models that used various asthma
biosignals and environmental factors. These predictive
models can help asthmatic patients predict asthma
attacks in advance, and thus preventive measures
can be taken. Fifteen different asthma attack
predictive models were selected for this review.
Asthma attack predictive models become more
significant when using both patient’s biosignal and
environmental factors. There is a lack of utilizing
advanced machine learning methods, like deep
learning techniques. Besides, there is a need to build
smart healthcare systems that provide patients with
decision-making systems to identify risk and visualize
high-risk regions.
There were four works [23, 25, 33, 36] that used real-time computing to predict
asthma attacks per individual. Different wireless sensors were employed to
acquire data from users and the environment. Hosseini et al. [23] developed a
model to divide the risk of having an asthma episode into three categories (low,
medium, and high risk). The biosignals data were recorded through a built-in
smartwatch wireless sensor, while the environmental data were acquired
from different meteorological wireless sensors. Data were analyzed in real-
time through the cloud platform.](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-70-2048.jpg)














![Asthma Phenotypes #3
Zhan et al. (2023): “Identification of cough-variant asthma
phenotypes based on clinical and pathophysiologic data”
Cough-variant asthma (CVA) is a subtype of asthma that usually
presents solely with cough without any other symptoms such as
dyspnea or wheezing [Corrao et al. 1979]. In cough-predominant
asthma cough is the most predominant symptom but other symptoms
are also present such as dyspnea and/or wheeze [Niimi 2008]. CVA may
respond differently to antiasthmatic treatment. There are limited data
on the heterogeneity of CVA. We aimed to classify patients with CVA
using cluster analysis based on clinicophysiologic parameters and to
unveil the underlying molecular pathways of these phenotypes with
transcriptomic data of sputum cells.
Cluster 1 was characterized by female predominance, late onset,
normal lung function, and a low proportion of complete resolution of
cough (60.8%) after antiasthmatic treatment. Patients in cluster 2
presented with young, nocturnal cough, atopy, high type 2
inflammation, and a high proportion of complete resolution of cough
(73.3%) with a highly upregulated coexpression gene network that
related to type 2 immunity. Patients in cluster 3 had high body mass
index, long disease duration, family history of asthma, low lung
function, and low proportion of complete resolution of cough (54.1%).](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-87-2048.jpg)





![Profiling for Biologics
van der Burg and Tufvesson (2023): “Is asthma's
heterogeneity too vast to use traditional phenotyping
for modern biologic therapies?”
Clinical trials of novel biologics and cluster analysis in asthma
still heavily rely on clinically-defined phenotypes. Biologics are
unique in that they target a specific antibody, molecule, or cell
involved in asthma. Because of this, they are known as
“precision” or “personalized” therapy. Bioprofiling patients with
asthma could provide more targetable groups for modern
biologics.
Quality-of-life questionnaire scores consistently show how
patients improve when they perceive to have less asthma
symptoms, sleep problems, anxiety and depression as well as
experience the regained capability to actively participate in
daily life. However, such reports of asthma do not include precise
measurements that reflect the underlying mechanisms of the
disease nor the severity of the airway inflammation in individual
patients. Meanwhile, clinical and real-life evidence show that a
substantial number of asthma patients appear to have relative
corticosteroid resistance and thus remain uncontrolled when
treated with the one-size-fits-all approach.
Unfortunately, to date, clinical trials investigating novel biologics in asthma heavily
rely on clinically defined phenotypes as target populations, often without a clear link
to the underlying biological profile (at treatment initiation nor over time) within these
target populations[[19],[20]]
. In the majority of such trials, T2-high severe asthma has
become the primary target population because of the availability of some
biomarkers that can be used to determine this phenotype[[16],[19]]
. Vice-versa, there is a
strong bias to developing T2 targeted biologics[[21]]
, leaving the T2-low and non-T2
mechanisms largely undefined and untargeted[[20]]
. Even within the T2-high asthma
phenotype landscape, the response to existing biologics is unpredictable and can be
associated with a substantial proportion of non-responders or partial responders[[22],
[23]]
.
There is a lot to be investigated in the context of biologically-based clustering of
asthma patients. Changes in the clinico-bioprofile must also be assessed, over time
with and without the intervention of biologics. There are also no clear indications that
current clustering techniques are stable within a patient over time nor did any multi-
center studies assess site-to-site specific effects on clusters. Future studies should
consider generating this data as it will become highly useful when we first begin to
form unbiased cluster groups of patients with asthma for selection of biologics both in
the context of clinical trials as well as in clinical practice.
By using longitudinal clinico-biological data from large asthma registries and
defining biologically-based clusters of asthma patients, we believe the development
and application of biologics will become more precise to asthma patients as
individuals.](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-94-2048.jpg)



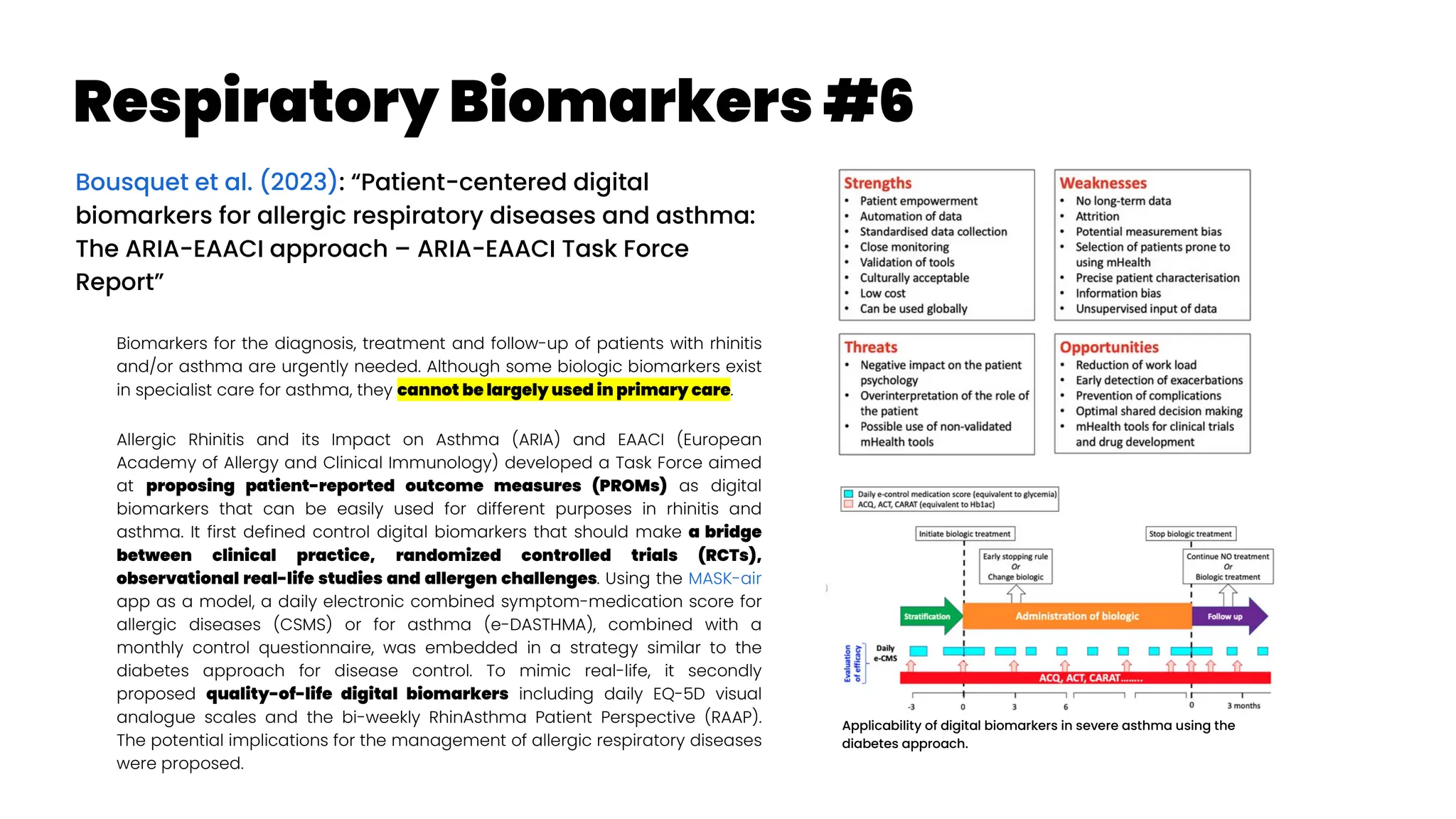
![Treatable Traits #1a
Gibson et al. (2023): “Treatable traits, combination inhaler
therapy and the future of asthma management” Cited by 1
Currently, inhaled corticosteroids (ICS), with or without long-acting beta
agonists (LABA), are the cornerstone of asthma management, and recently
international guidelines recognized the importance of combination inhaler
therapy (ICS/LABA) even in mild asthma. In future, ultra-long-acting
personalized medications and smart inhalers will complement combination
inhaler therapy in order to effectively addresses issues such as adherence,
inhaler technique and polypharmacy (both of drugs and devices). Asthma is
now acknowledged as a multifaceted cluster of disorders and the treatment
model has evolved from one-size-fits-all to precision medicine approaches
such as treatable traits (McDonald et al. 2019, Thomas 2022
; TTs, defined as measurable
and treatable clinically important factors) which encourages the quality use
of medications and identification and management of all underlying
behavioural and biological treatable risk factors.
TT requires research and validation in a clinical context and the
implementation strategies and efficacy in various settings
(primary/secondary/tertiary care, low-middle income countries) and
populations (mild/moderate/severe asthma) are currently evolving.
Combination inhaler therapy and the TTs approach are complementary
treatment approaches. This review examines the current status of personalized
medicine and combination inhaler therapy, and describes futuristic views for
these two strategies.
What is a treatable trait? An identifiable feature that can be assessed
and targeted by treatment to improve a clinical outcome is called a
treatable trait.20
Three characteristics are present in each treatable trait
10, 21, 22
: (1) clinical significance (i.e., the trait is associated with a clinical
outcome, for example, asthma exacerbation, quality of life [QoL] or
asthma control); (2) detectable (i.e., measurable via specific and
validated ‘trait identification markers [TIM]’, e.g., the trait of T2
inflammation is assessed by the TIM of circulating eosinophils); and,
(3) treatable (an effective treatment is available).
Eosinophilic/T2 airway inflammation is an excellent example of a
treatable trait in asthma. This endotype is mediated by specific cytokines
such as interleukin (IL)-5.23
The levels of eosinophilic inflammation are
directly proportional to exacerbation risk indicating its clinical relevance.
Moreover, it can be detected via blood eosinophil count (a TIM) and can
be treated via corticosteroids (inhaled or oral) or T2-targeted biologics.
Hence, eosinophilic airway inflammation meets all the criteria for a
treatable trait (i.e., relevant, detectable and treatable).
Treatable traits approach has already been described for use in tertiary
care settings. The future development of treatable traits will identify how
to adapt treatable traits to different settings, such as primary care.
Lung sounds / other biomarkers the (2) of treatable traits (“detectable”)](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-99-2048.jpg)











![Asthma Comorbidities
Kwon et al. (2021): “Risk, Mechanisms and Implications of Asthma-
Associated Infectious and Inflammatory Multimorbidities (AIMs)
among Individuals With Asthma: a Systematic Review and a Case
Study” Cited by 5
Our prior work and the work of others have demonstrated that asthma
increases the risk of a broad range of both respiratory (e.g., pneumonia and
pertussis) and non-respiratory (e.g., zoster and appendicitis) infectious
diseases as well as inflammatory diseases (e.g., celiac disease and
myocardial infarction [MI]), suggesting the systemic disease nature of
asthma and its impact beyond the airways. We call these conditions
asthma-associated infectious and inflammatory multimorbidities
(AIMs). At present, little is known about why some people with asthma are
at high-risk of AIMs, and others are not, to the extent to which controlling
asthma reduces the risk of AIMs and which specific therapies mitigate
the risk of AIMs. These questions represent a significant knowledge gap in
asthma research and unmet needs in asthma care, because there are no
guidelines addressing the identification and management of AIMs.
This is a systematic review on the association of asthma with the risk of AIMs
and a case study to highlight that 1) AIMs are relatively under-recognized
conditions, but pose major health threats to people with asthma; 2) AIMs
provide insights into immunological and clinical features of asthma as a
systemic inflammatory disease beyond a solely chronic airway disease; and
3) it is time to recognize AIMs as a distinctive asthma phenotype in order
to advance asthma research and improve asthma care. An improved
understanding of AIMs and their underlying mechanisms will bring valuable
and new perspectives improving the practice, research, and public health
related to asthma.](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-111-2048.jpg)

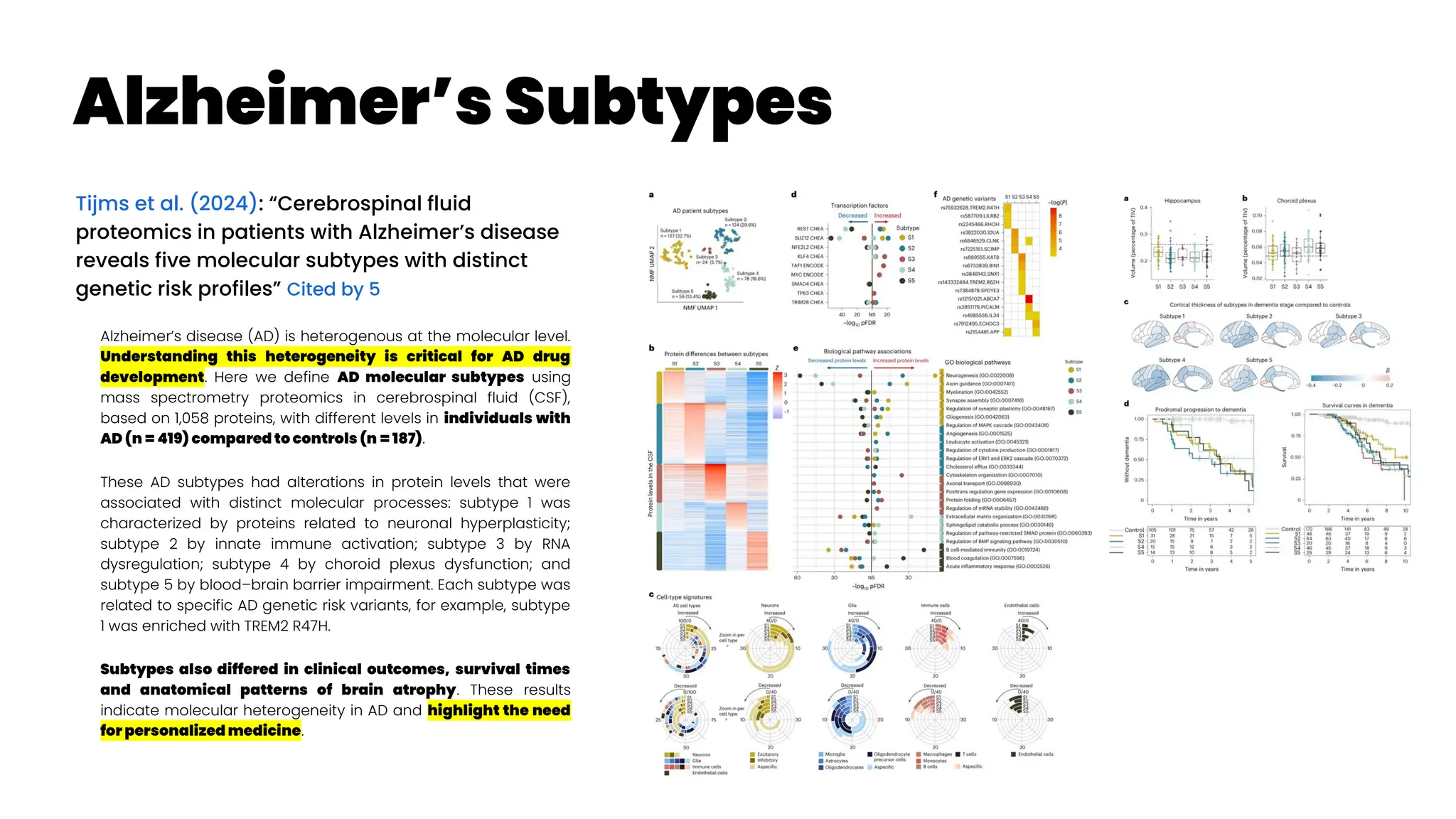


![Precision Drug Development for Alzheimer’sc
Cheng et al. (2024): “Artificial intelligence and open
science in discovery of disease-modifying medicines for
Alzheimer’s disease”
A recent study presented network-based disease-progression-
specific drug repurposing for AD based on neuroimaging-derived
disease stages (Savva et al. 2022)
. Using three types of PET brain imaging
for microglial activation ([11C]PBR28)
, amyloid-b (Ab) ([18F]AZD4694)
, and tau
([18F]MK-6240)
, a recent study showed that the co-occurrence of Ab,
tau, and microglia abnormalities was the strongest predictor of
cognitive impairment (Pascoal et al. 2021)
.
This study highlighted that synergistically targeting microglial
abnormality, tau, and Ab endophenotypes could offer more
clinical benefits compared to targeting each endophenotypes
alone, supporting previous endophenotype-based drug
repurposing in AD (Fang et al. 2021)
.
Combination therapies via targeting molecular networks or
pathways derived from multi-omics data from individuals with well-
characterized proteinopathies and microglia abnormalities may
offer more effective treatment approaches compared to
monotherapies targeting one protein/microglia subtype. More
details about AI-based or network-based drug combination design
can be found in recent studies (94–96).
As drug development is a complex process involving many
steps, multi-modal machine learning tools can
significantly reduce the time and cost of drug
development. For instance, multi-modal machine learning
approaches (Venugopalan et al. 2021)
improve accuracy of patient
subphenotyping during clinical trial design by
assembling neuroimaging, genetic, and multi-omics
profiling data. With the help of deep learning, effective
representations can be learned for different data
modalities (Zhavoronkov et al. 2019)
, which can then be fused by
simple concatenation or more complicated nonlinear
transformation113 to perform downstream tasks such as
molecular design, PK and BBB property evaluation and
optimization, and robotics-based chemical synthesis,
which can greatly accelerate the drug discovery and
development process. If broadly applied, AI-based tools
will accelerate the development of disease-modifying
treatments for AD.](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-116-2048.jpg)





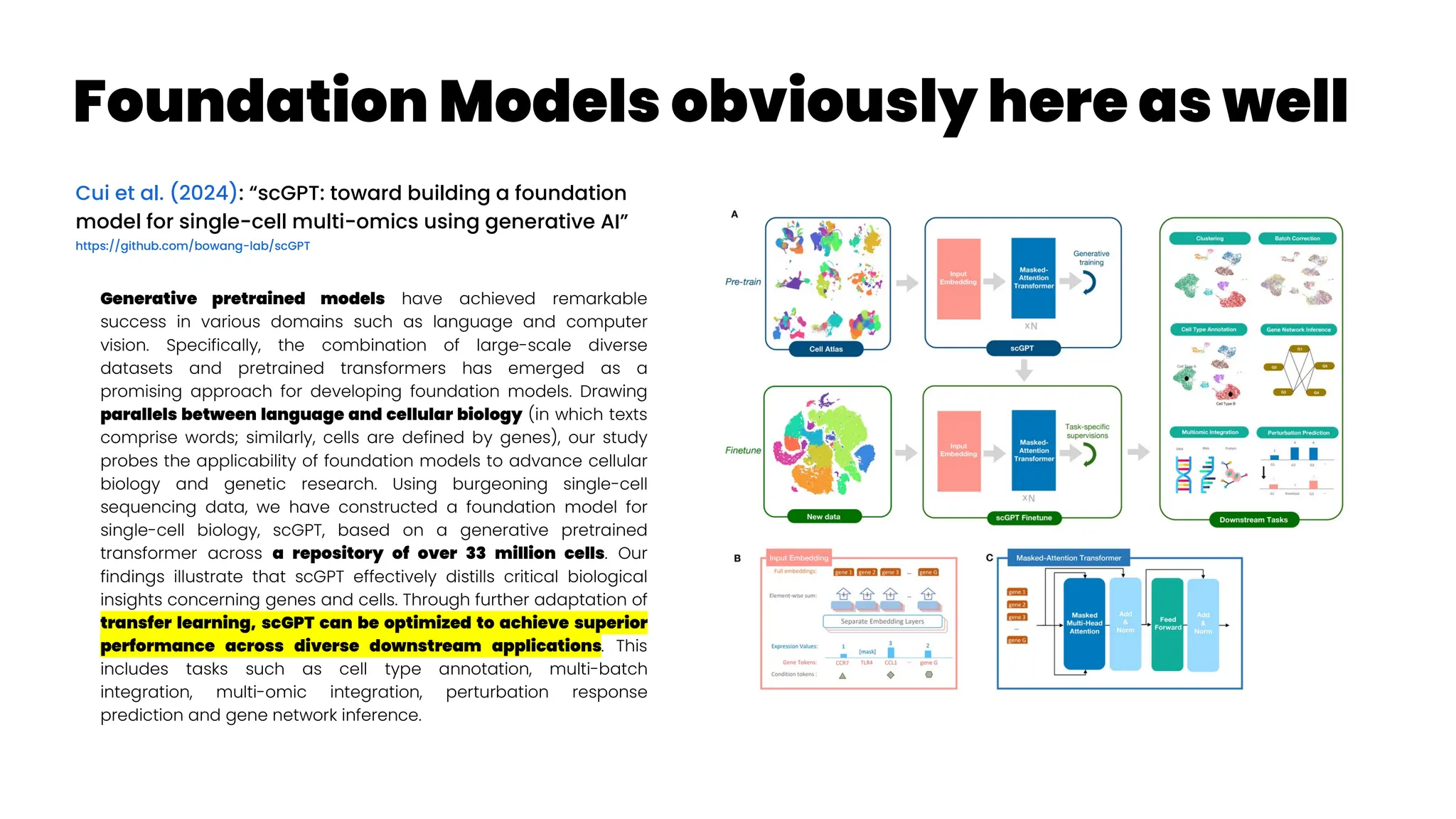








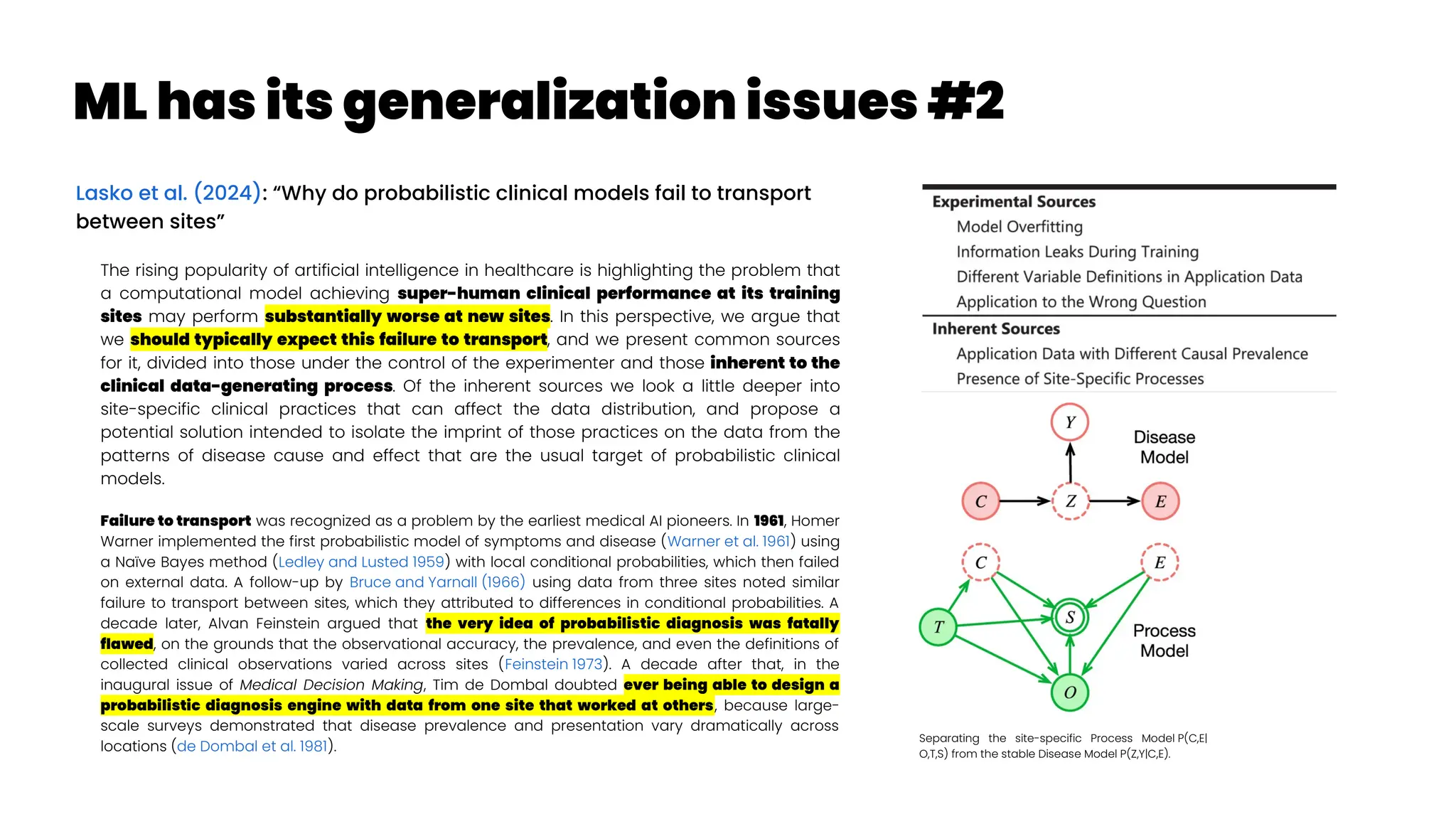
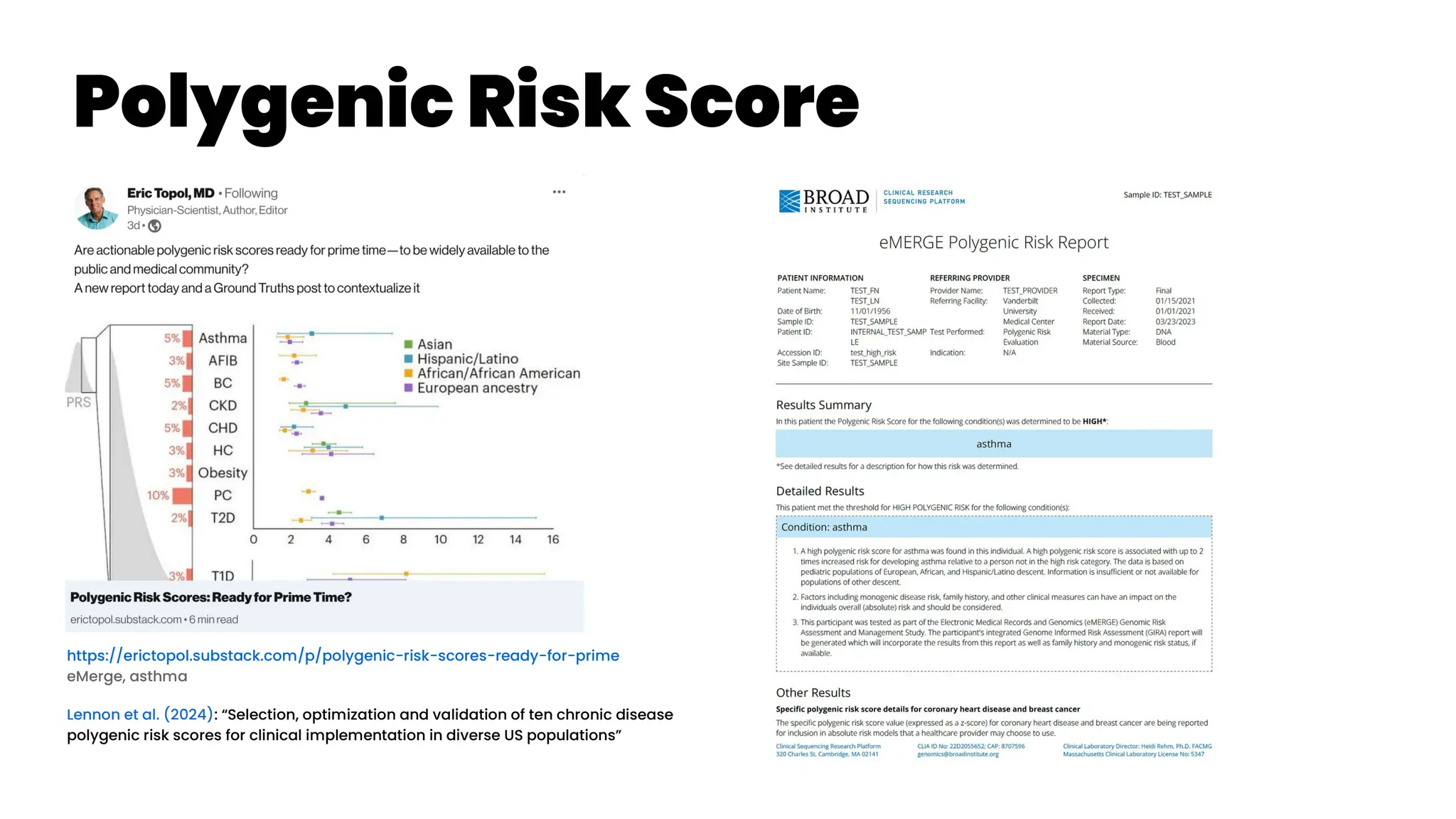

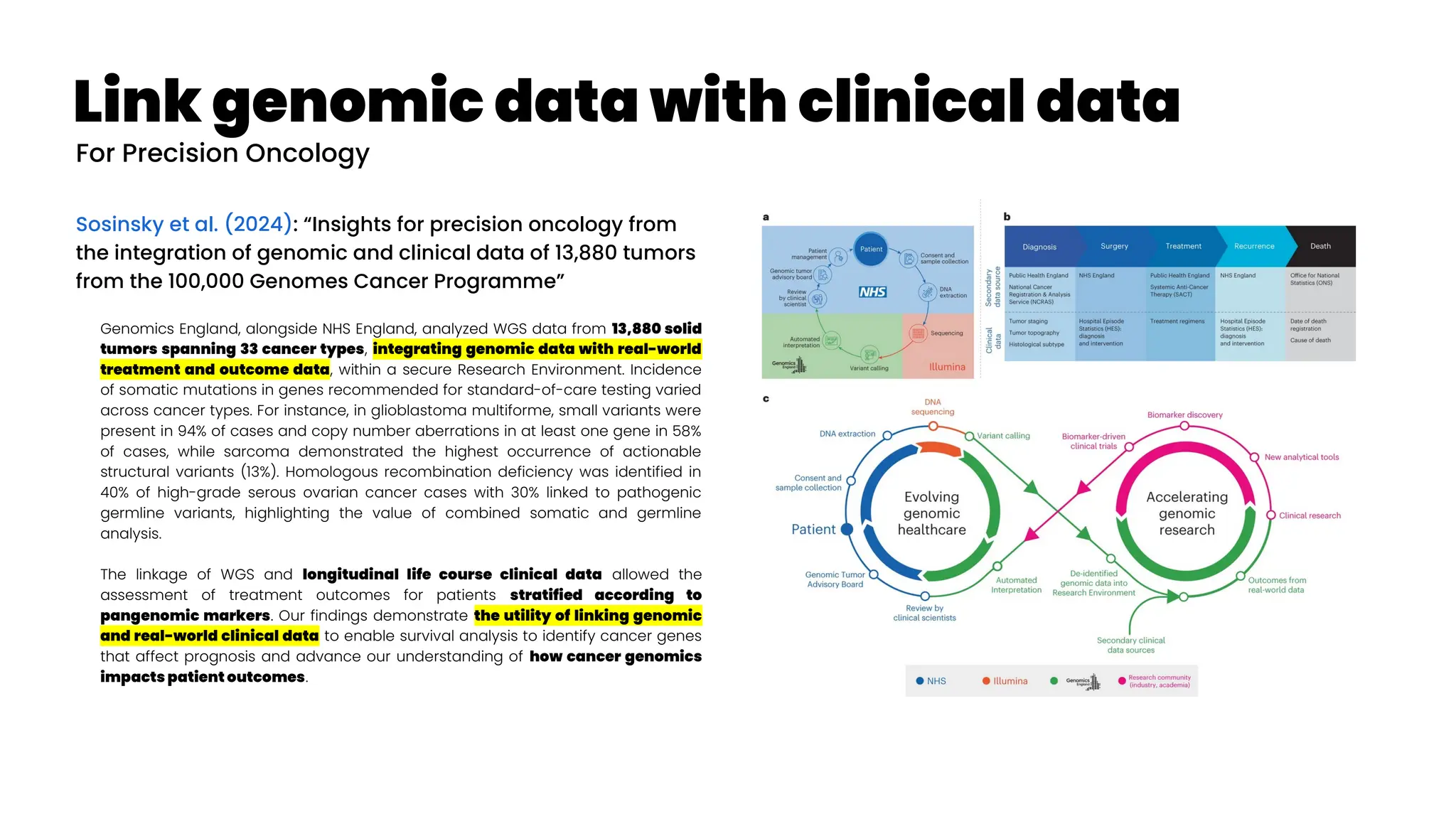




![Asthma Management Tool
Zheng et al. (2024): “Clinical Needs Assessment of a Machine
Learning–Based Asthma Management Tool: User-Centered
Design Approach”
Personalized asthma management depends on a clinician’s ability
to efficiently review patient’s data and make timely clinical
decisions. While machine learning (ML) and clinical decision support
tools are well-positioned as potential solutions, the translation of such
frameworks requires that barriers to implementation be addressed in
the formative research stages.
The : Asthma Guidance and Prediction System (A-GPS) tool is an ML-
based CDS tool accessible from “within” the EHR workflow. It aims to
summarize all asthma-related context information extracted from the
EHR on 1 screen page [Seol et al. 2020, Seol et al. 2021]. The tool will be
embedded with a functional component of the asthma exacerbation
(AE) risk model, which applies ML algorithms to predict a patient’s
risk of exacerbation in 1 year [Overgaard et al. 2022]. Moreover, the
reason for the development of the AE risk model in this context was to
provide supplemental information to improve the care of patients with
asthma, not replace the expertise of clinicians.](https://image.slidesharecdn.com/wearablemicprecision-240326094528-3961c2da/75/Precision-Medicine-for-personalized-treatment-of-asthma-139-2048.jpg)




