Embed presentation
Download as PDF, PPTX






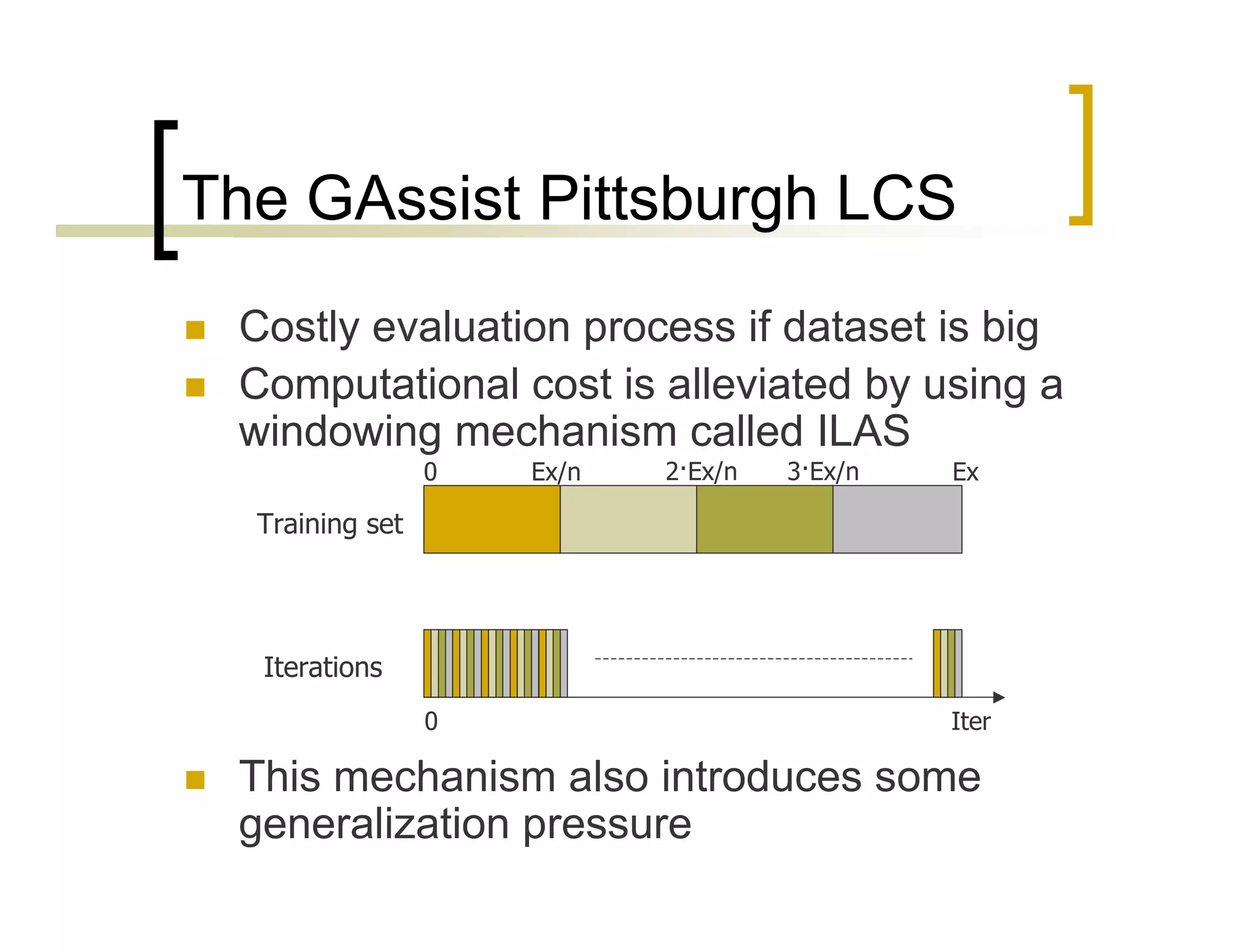
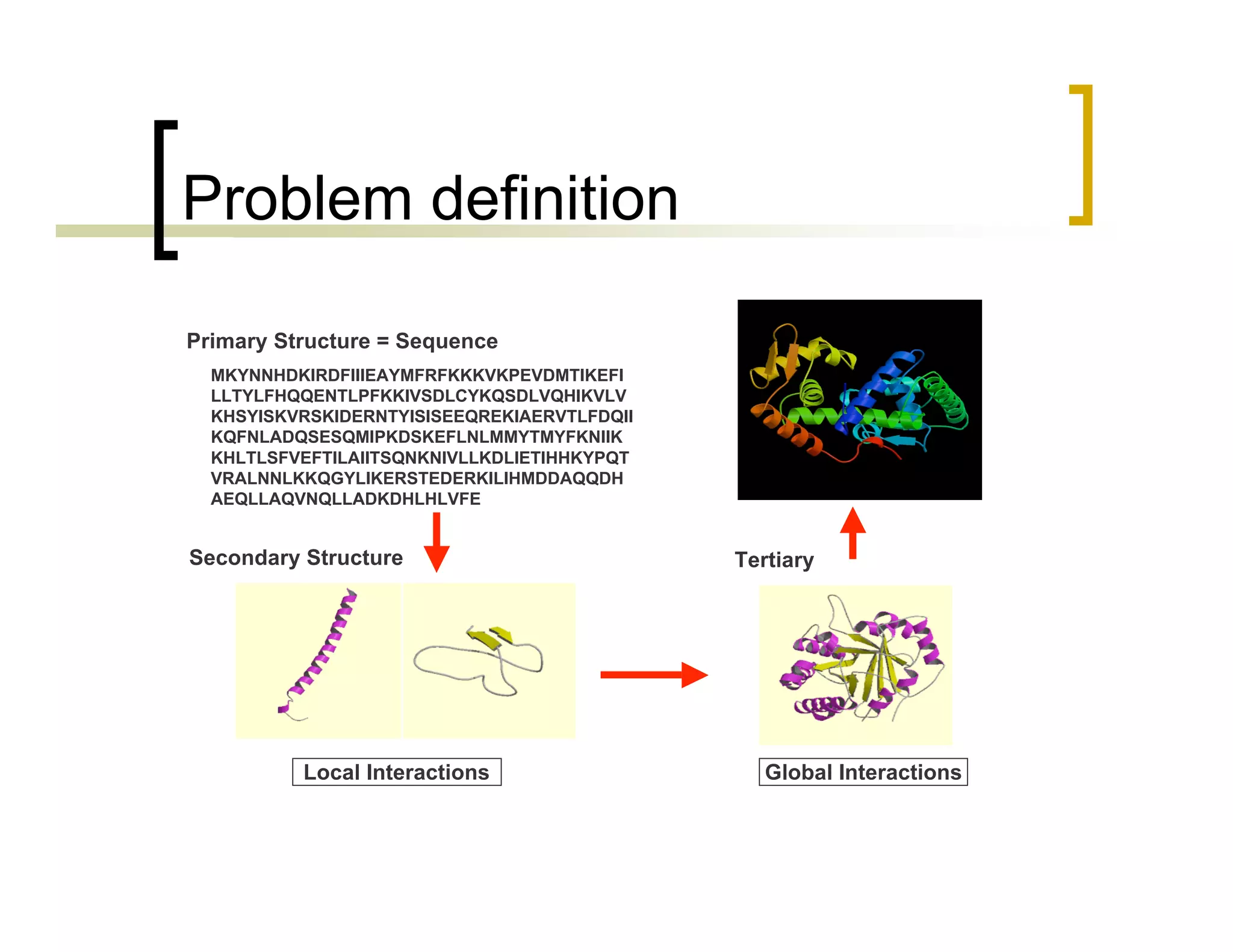
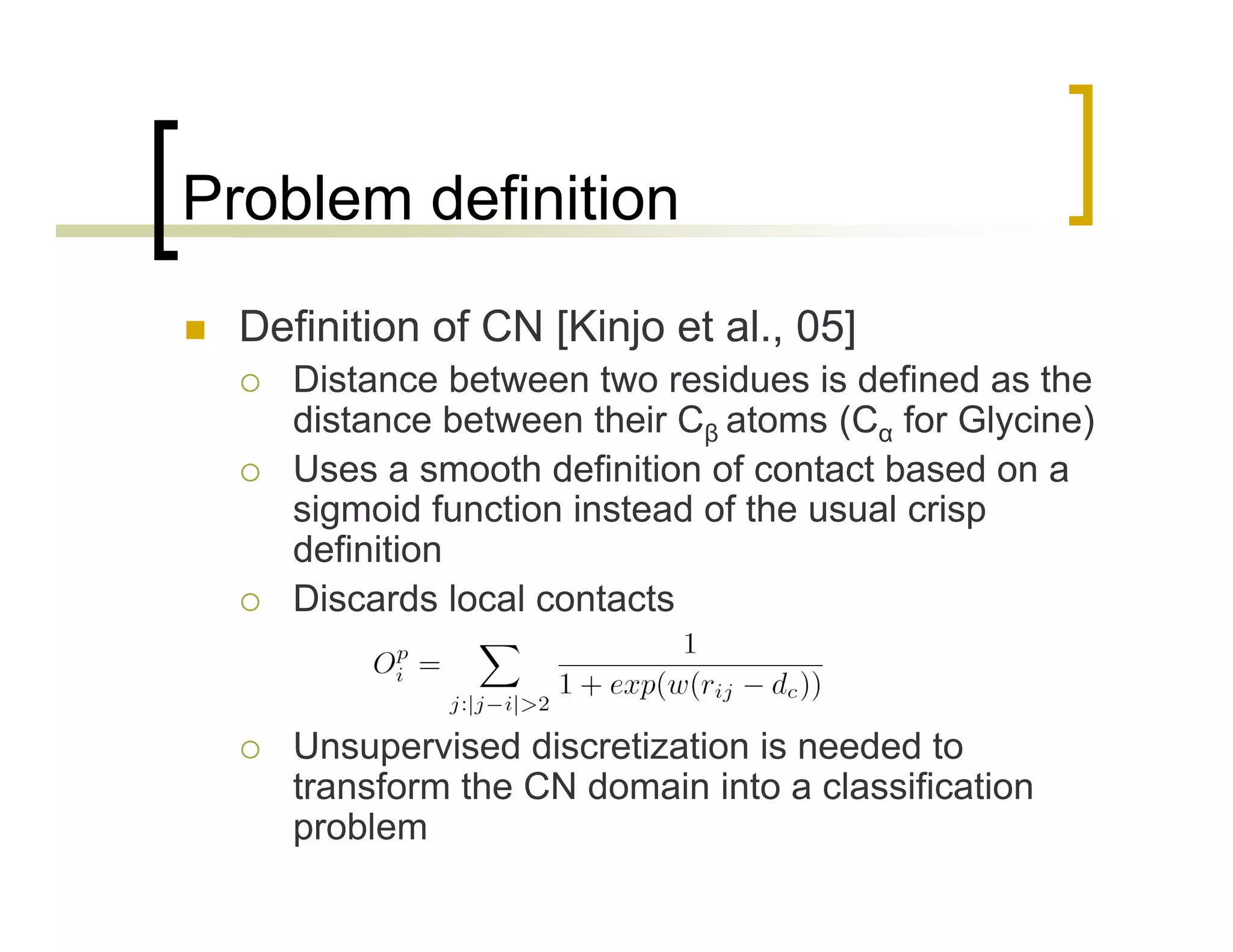

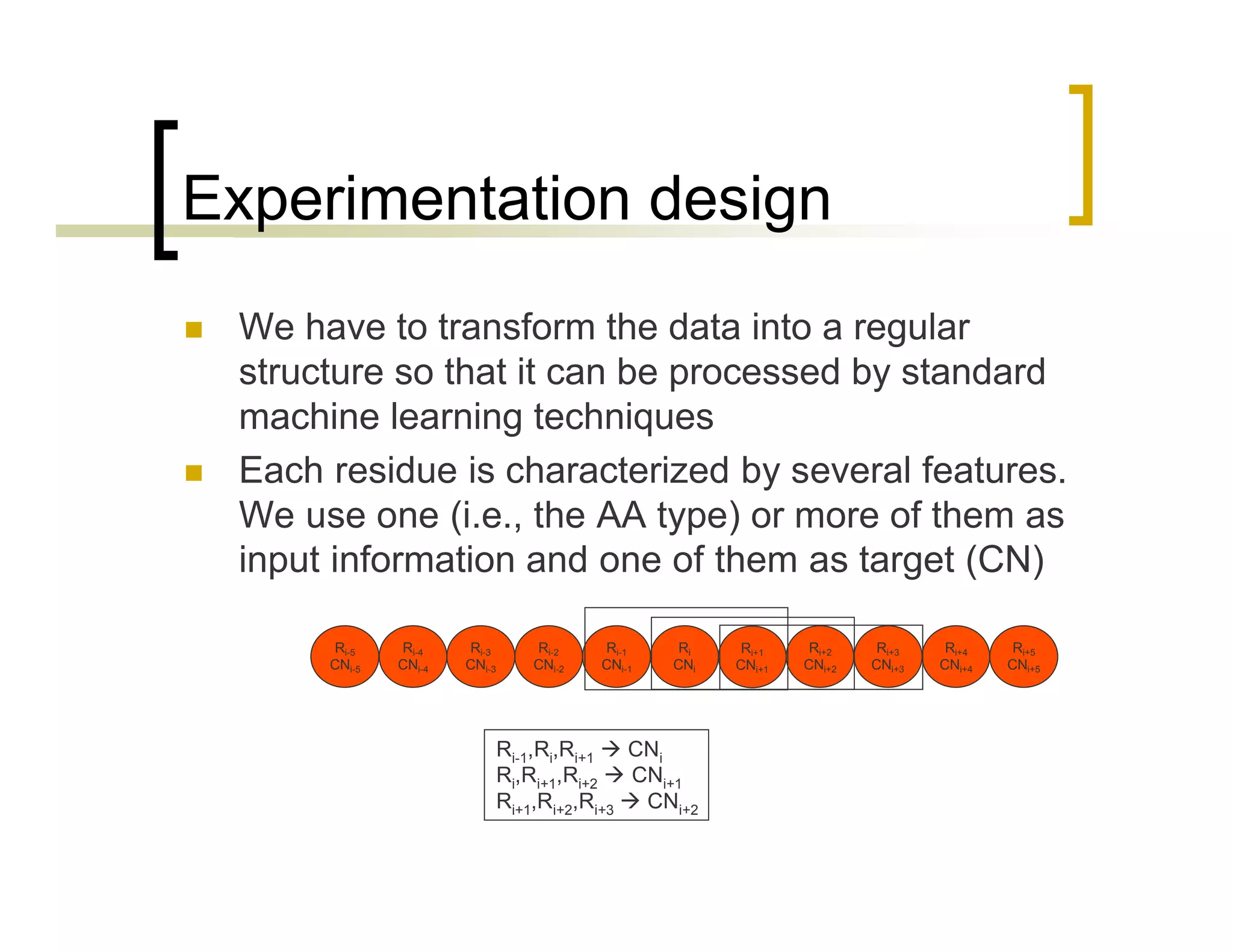
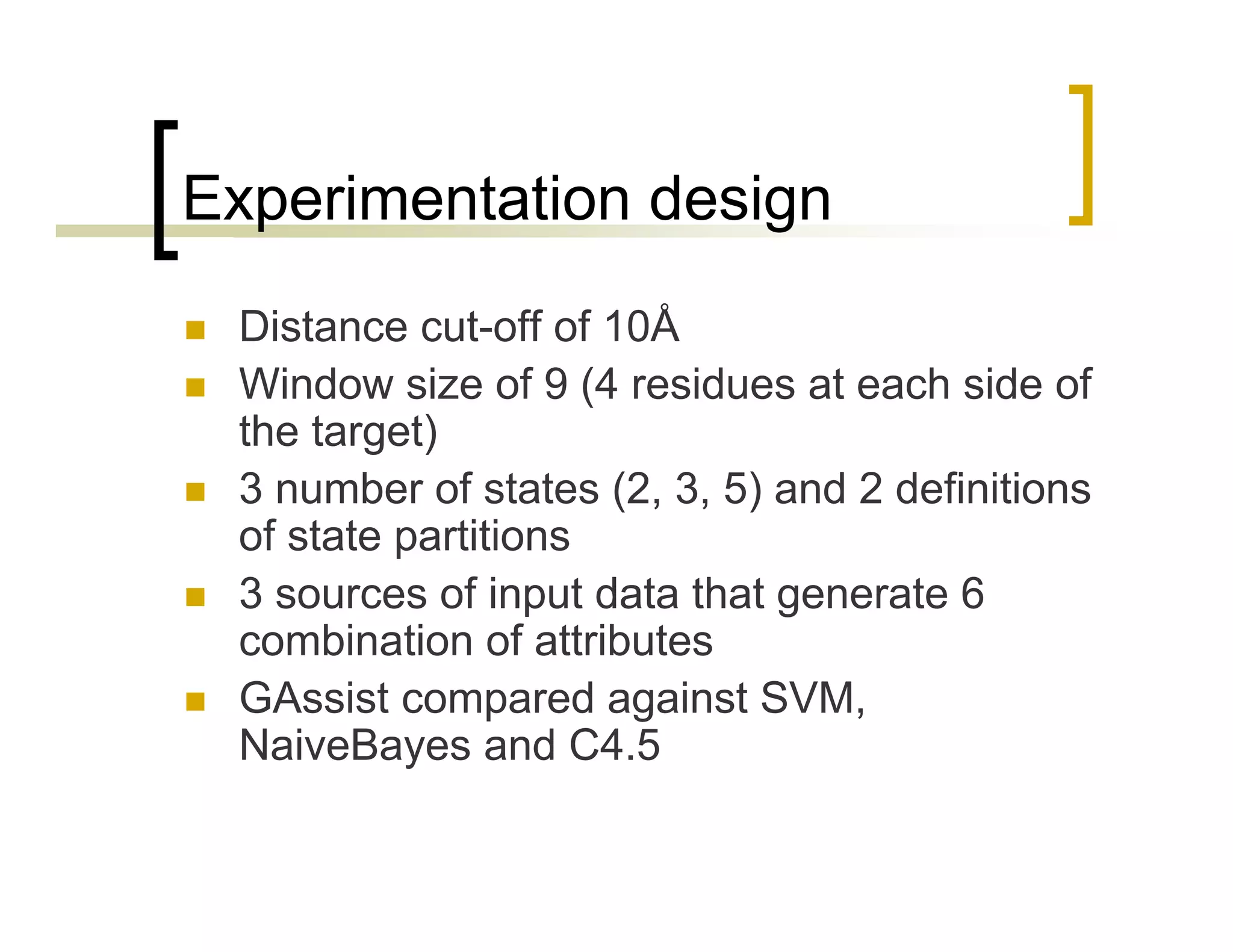
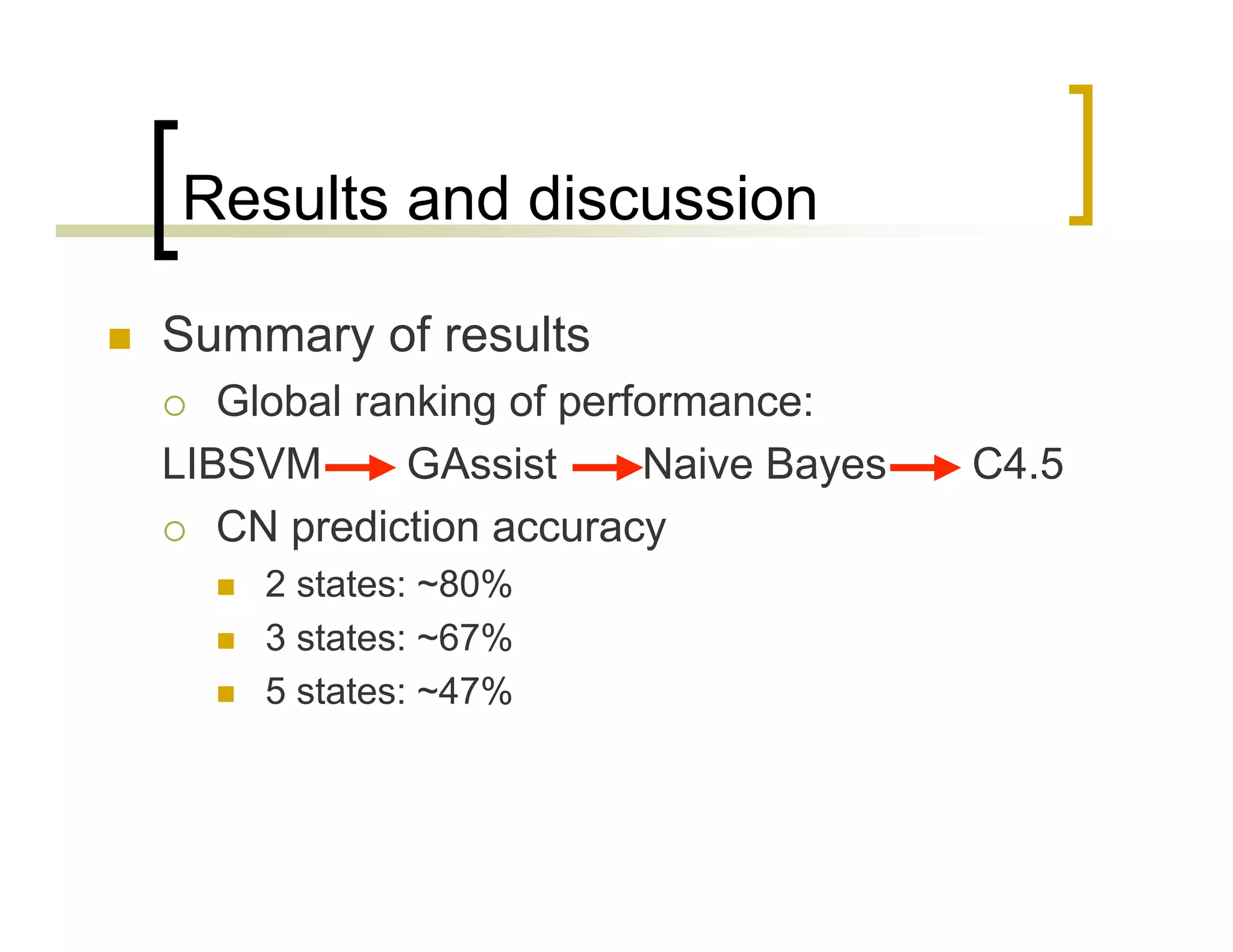

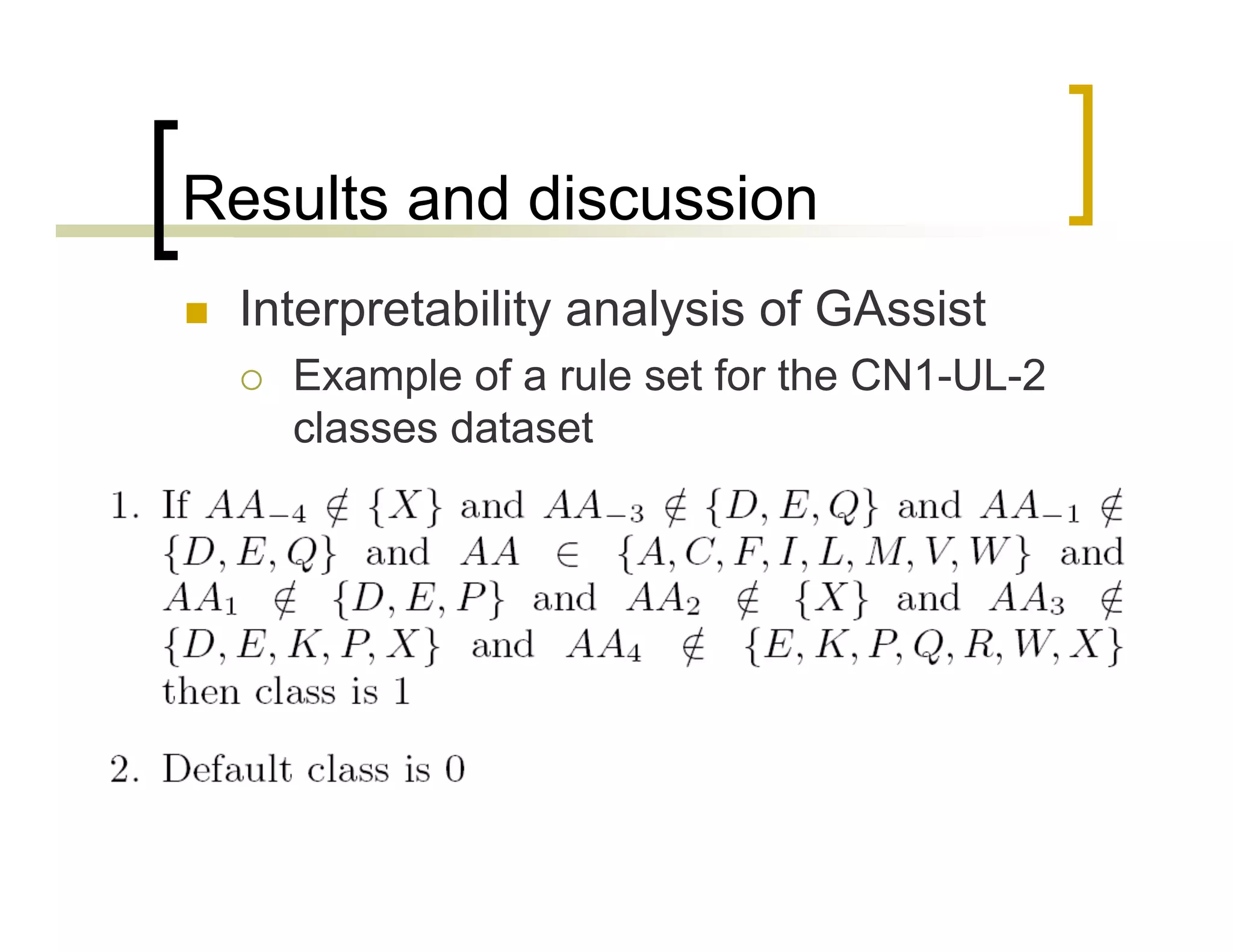
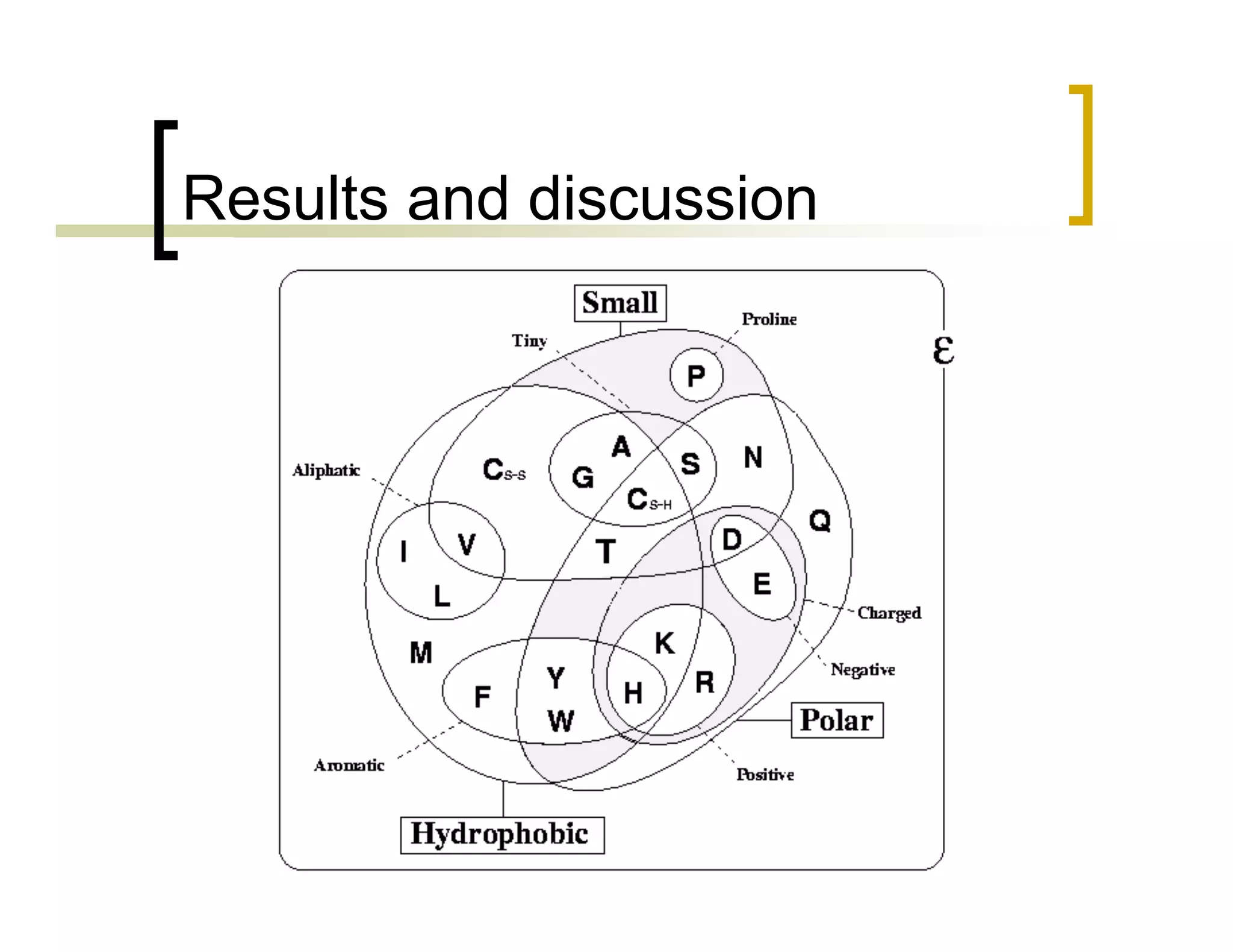

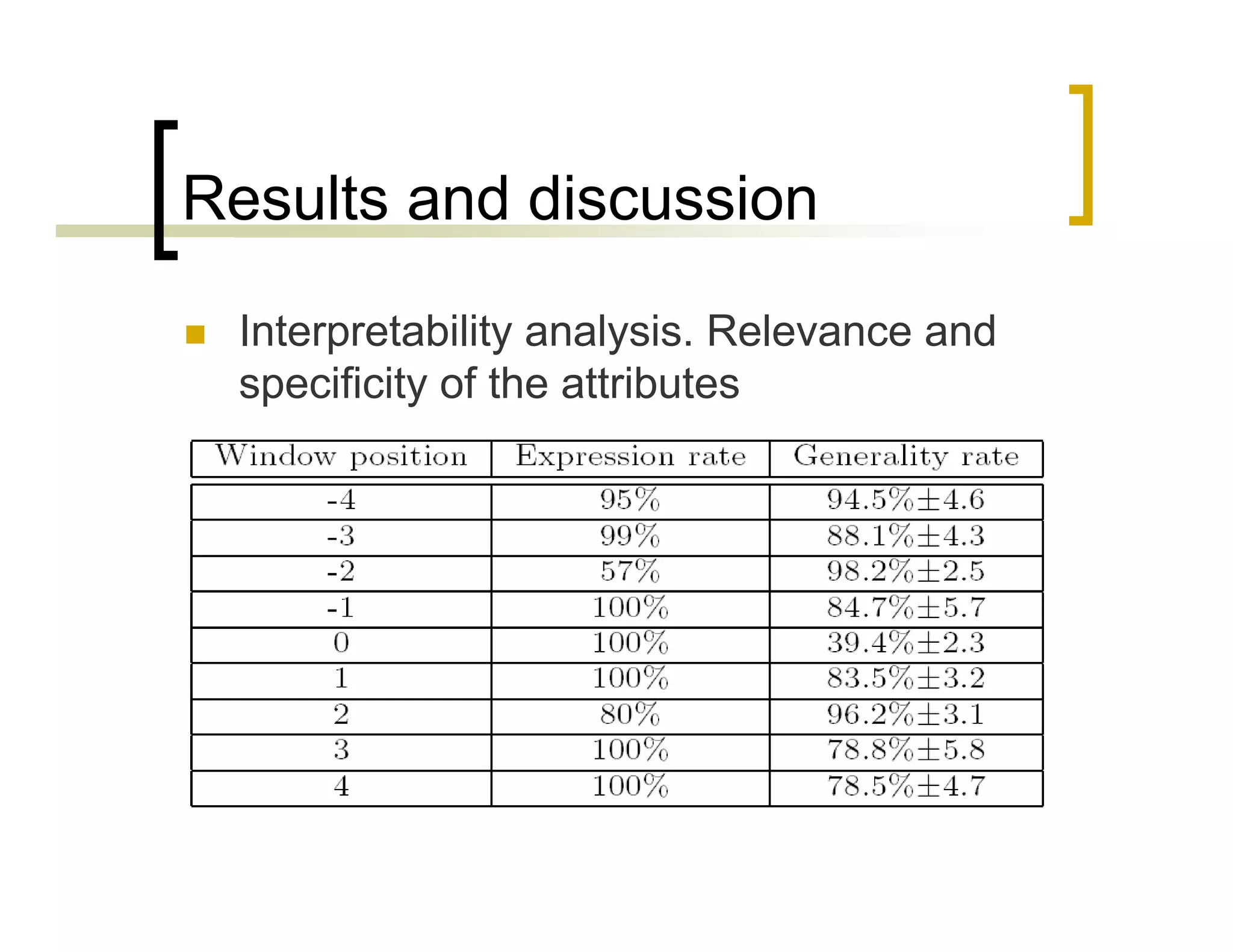
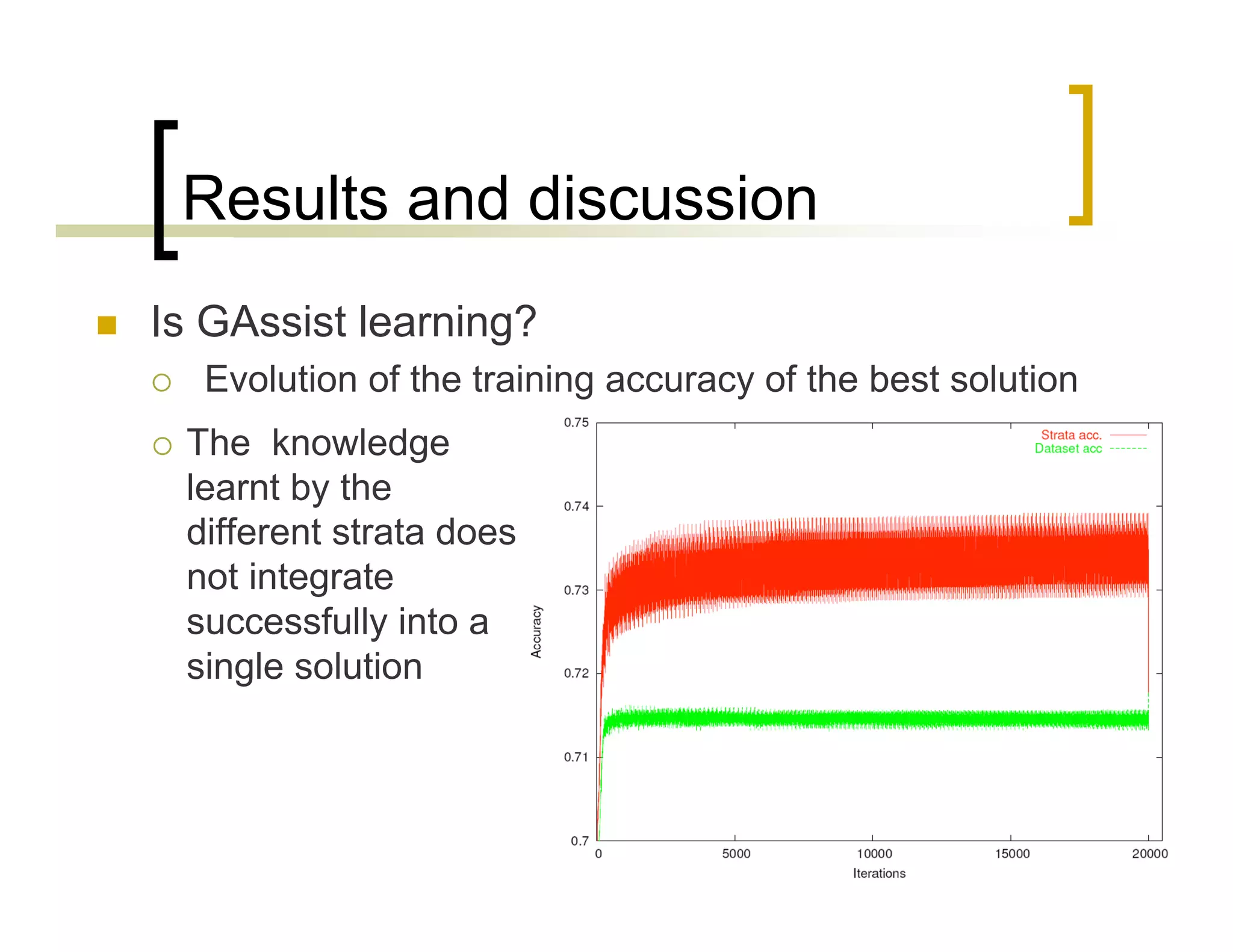




This document summarizes research using a Pittsburgh Learning Classifier System (LCS) called GAssist to predict protein structure by determining coordination numbers (CN). The researchers tested GAssist on a dataset of over 250,000 protein residues, comparing it to support vector machines, Naive Bayes, and C4.5 decision trees. While support vector machines achieved the best accuracy, GAssist produced more interpretable and compact rule sets at the cost of lower performance. The researchers analyzed the interpretability and scalability of GAssist for this challenging bioinformatics problem, identifying avenues for improving its accuracy while maintaining explanatory power.

























