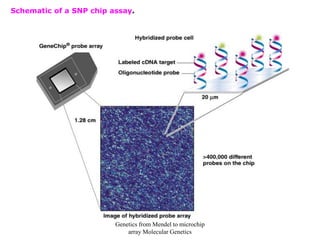
This document discusses molecular genetic methods such as polymerase chain reaction (PCR), DNA sequencing, DNA fingerprinting, and single nucleotide polymorphisms. It provides details on how each method works, including how PCR amplifies DNA, the process of manual and automated DNA sequencing, using variable number tandem repeats as markers for DNA fingerprinting, and applications of these molecular genetic techniques.