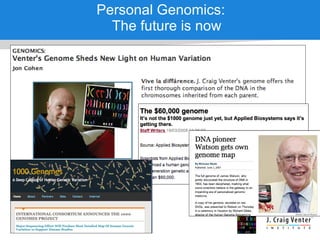
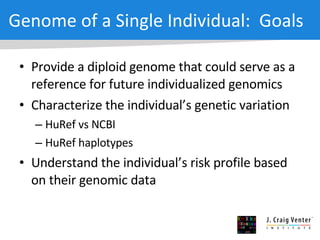
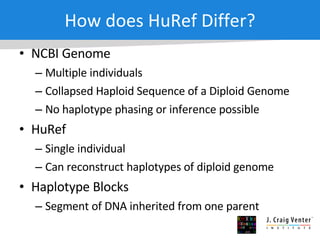
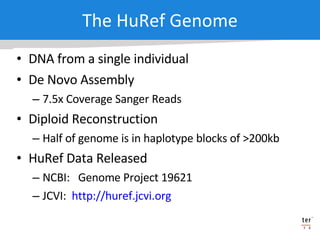
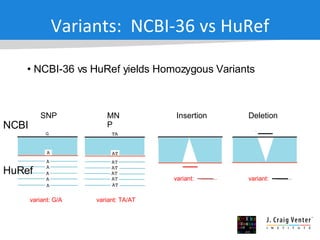
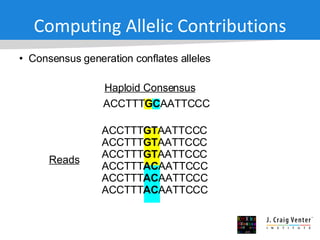
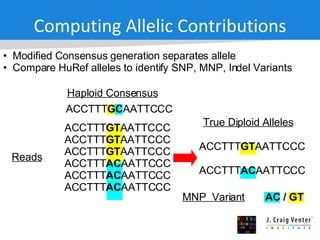
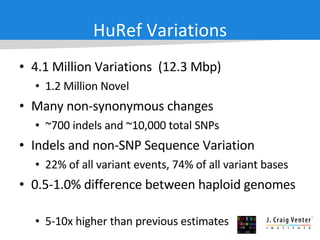
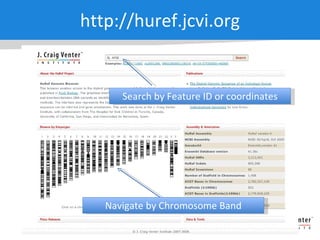
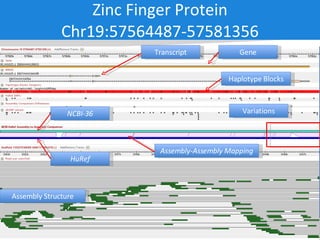
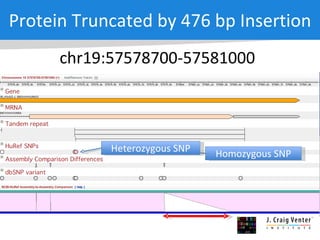
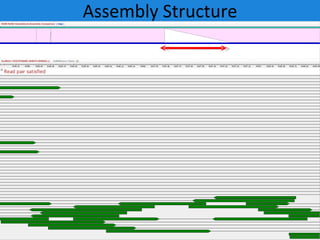
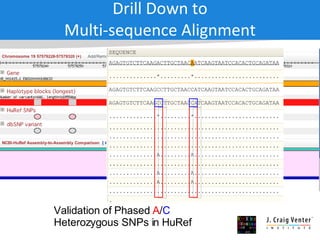
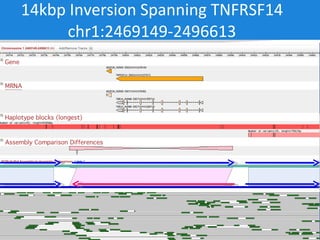
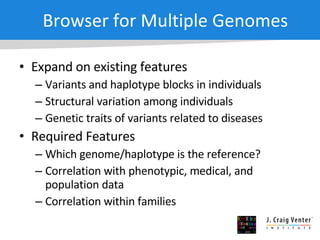
The document discusses the Huref project, which aims to provide a comprehensive diploid genome for an individual, serving as a reference for future individualized genomics and characterizing genetic variations. It highlights the differences between the Huref browser and existing databases, emphasizing its ability to reconstruct haplotypes and offer high-performance tools for genetic analysis. The document concludes with the potential impact of Huref on research and the challenges posed by the growing data volumes in genomics.





![Conclusion A high performance visualization tool for an individual genome Validation of variants Comparison with NCBI-36 Planned extensions for multi-genome era Website: http://huref.jcvi.org Contact: [email_address]](https://image.slidesharecdn.com/hurefbioitworld2008final-1213385186104705-9/85/Human-Reference-Genome-Browser-Presentation-at-BIO-ITWorld-2008-22-320.jpg)
