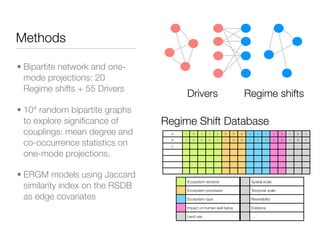
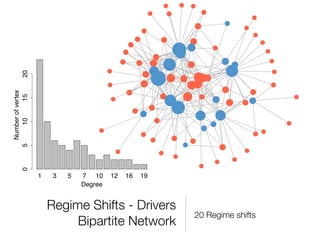
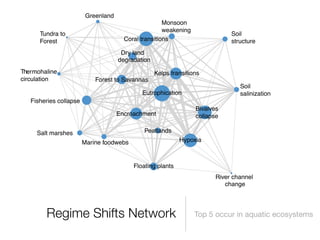
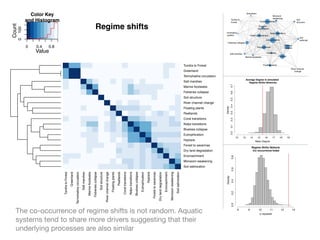
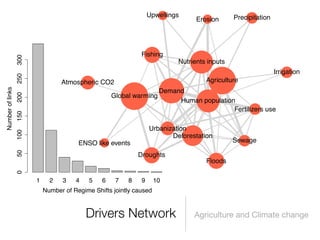
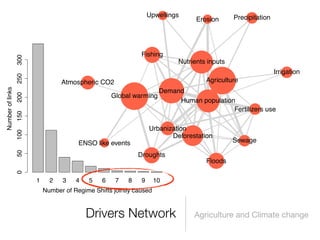
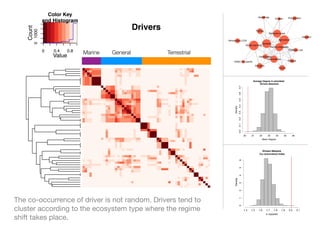
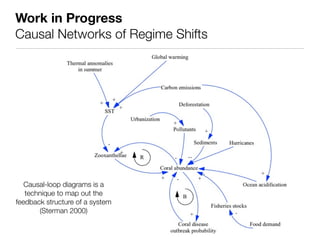
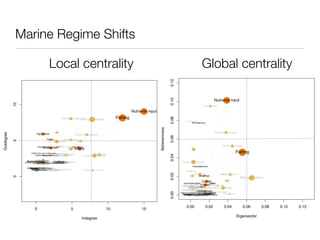
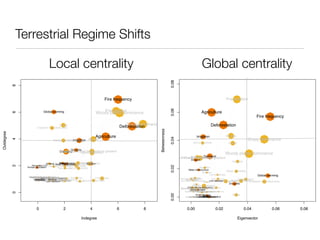
The document discusses two networks constructed from genetic disorder and disease gene data: the human disease network (HDN) and the disease gene network (DGN). The HDN connects disorders that share disease genes, revealing many connections between disorders. The DGN connects disease genes associated with the same disorders. Most disorders relate to a few genes, while a few disorders like cancers relate to dozens of genes. The networks provide an integrated framework to explore relationships between genetic disorders and disease genes.



![The human disease network Network Properties of Complex Human Disease Genes
Kwang-Il Goh*†‡§, Michael E. Cusick†‡¶, David Valleʈ, Barton Childsʈ, Marc Vidal†‡¶**, ´ ´
and Albert-Laszlo ´
Barabasi*†‡**
*Center for Complex Network Research and Department of Physics, University of Notre Dame, Notre Dame, IN 46556; †Center for Cancer Systems Biology
Identified through Genome-Wide Association Studies
(CCSB) and ¶Department of Cancer Biology, Dana–Farber Cancer Institute, 44 Binney Street, Boston, MA 02115; ‡Department of Genetics, Harvard Medical
School, 77 Avenue Louis Pasteur, Boston, MA 02115; §Department of Physics, Korea University, Seoul 136-713, Korea; and ʈDepartment of Pediatrics and the Fredrik Barrenas1.*, Sreenivas Chavali1., Petter Holme2,3, Reza Mobini1, Mikael Benson1
McKusick–Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205
˚ ˚
1 The Unit for Clinical Systems Biology, University of Gothenburg, Gothenburg, Sweden, 2 Department of Physics, Umea University, Umea, Sweden, 3 Department of
Edited by H. Eugene Stanley, Boston University, Boston, MA, and approved April 3, 2007 (received for review February 14, 2007) Energy Science, Sungkyunkwan University, Suwon, Korea
A network of disorders and disease genes linked by known disorder– known genetic disorders, whereas the other set corresponds to all
gene associations offers a platform to explore in a single graph- known disease genes in the human genome (Fig. 1). A disorder and
theoretic framework all known phenotype and disease gene associ- a gene are then connected by a link if mutations in that gene are Abstract
ations, indicating the common genetic origin of many diseases. Genes implicated in that disorder. The list of disorders, disease genes, and
DISEASOME
associated with similar disorders show both higher likelihood of associations between them was obtained from the Online Mende- Background: Previous studies of network properties of human disease genes have mainly focused on monogenic diseases
physical interactions between their products and higher expression lian Inheritance in Man (OMIM; ref. 18), a compendium of human or cancers and have suffered from discovery bias. Here we investigated the network properties of complex disease genes
profiling similarity for their transcripts, supporting the existence of disease genes and phenotypes. As of December 2005, this list identified by genome-wide association studies (GWAs), thereby eliminating discovery bias.
distinct disease-specific functional modules. Wedisease phenome find that essential disease genome
contained 1,284 disorders and 1,777 disease genes. OMIM initially
Human Disease Network
human genes are likely to encode hub proteins and are expressed Disease Gene Network Principal findings: We derived a network of complex diseases (n = 54) and complex disease genes (n = 349) to explore the
Ataxia-telangiectasia
focused on AR monogenic disorders but in recent years has expanded
(HDN)
widely in most tissues. This suggests that disease genes hypospadias Perineal also would
Androgen insensitivity
(DGN)
to include complex traits and the associated genetic mutations that
ATM
shared genetic architecture of complex diseases. We evaluated the centrality measures of complex disease genes in
play a central role in the human interactome. In contrast, we find that confer susceptibility to these common disorders (18). Although this comparison with essential and monogenic disease genes in the human interactome. The complex disease network showed
T-cell lymphoblastic leukemia
the vast majority of disease genes are nonessential and show no
Charcot-Marie-Tooth disease
Papillary serous carcinoma
BRCA1
history introduces some biases, LMNA the disease gene record is far
and HEXB that diseases belonging to the same disease class do not always share common disease genes. A possible explanation could
tendency to encode hubLipodystrophy and their expression pattern indi-
proteins, BRCA2
from complete, OMIM represents the most complete and up-to-
ALS2
be that the variants with higher minor allele frequency and larger effect size identified using GWAs constitute disjoint parts
APPLIED PHYSICAL
Prostate cancer
cates that ataxia/paraplegia localized paraplegia syndrome
Spastic
they are Silver spastic in the functional periphery of the CDH1 BSCL2
date repository of all known VAPBdisease genes and the disorders they of the allelic spectra of similar complex diseases. The complex disease gene network showed high modularity with the size
SCIENCES
network. A selection-basedSandhoff disease model explains the observed difference
Ovarian cancer GARS
Amyotrophic lateral sclerosis confer. We manually classified each disorder into one of 22 disorder
GARS
of the largest component being smaller than expected from a randomized null-model. This is consistent with limited sharing
between essential and disease genes and also suggests that diseases
Lymphoma classes based on the physiological system affected [see supporting
HEXB
of genes between diseases. Complex disease genes are less central than the essential and monogenic disease genes in the
caused by somatic mutations should not be peripheral, a prediction
Spinal muscular atrophy
information (SI) Text, SI Fig. 5, and SI Table 1 for details].
KRAS AR
human interactome. Genes associated with the same disease, compared to genes associated with different diseases, more
we confirm for cancer genes. insensitivity
Androgen
Prostate cancer
Breast cancer Starting LMNA the diseasome bipartite graph we generated two
from ATM often tend to share a protein-protein interaction and a Gene Ontology Biological Process.
Perineal hypospadias
biological networks ͉ complex networks ͉ human genetics Pancreatic cancer
͉ systems
biologically relevant network projections (Fig. 1). In the ‘‘human
MSH2 BRIP1
BRCA2
biology ͉ diseasome Lymphoma
disease network’’ (HDN) nodes represent disorders, and two
PIK3CA BRCA1 Conclusions: This indicates that network neighbors of known disease genes form an important class of candidates for
Wilms tumor
Breast cancer
Wilms tumor
disorders are connected to each otherKRAS
TP53 RAD54L
if they share at least one gene identifying novel genes for the same disease.
Ovarian cancer
in which mutations are associated with both TP53 disorders (Figs. 1 and
D
Spinal muscular atrophy
ecades-long efforts to map human disease loci,Sandhoff disease
Pancreatic cancer at first genet- MAD1L1
2a). In the ‘‘disease gene network’’MAD1L1
(DGN) nodes represent disease
ically and later physically (1), followed by recent positional
Fanconi anemia
Papillary serous carcinoma
Lipodystrophy two
CHEK2
genes, andRAD54L genes are connected if they are associated with the Citation: Barrenas F, Chavali S, Holme P, Mobini R, Benson M (2009) Network Properties of Complex Human Disease Genes Identified through Genome-Wide
cloning of many disease T-cell lymphoblastic and genome-wide association
genes (2) leukemia Charcot-Marie-Tooth disease
PIK3CA
same disorder (Figs. 1 and 2b). Next, we discuss the potential of
VAPB Association Studies. PLoS ONE 4(11): e8090. doi:10.1371/journal.pone.0008090
studies (3), have generated an impressive list of disorder–gene
Ataxia-telangiectasia
these networks to help us understand and MSH2
CHEK2 CDH1
represent in a single Editor: Thomas Mailund, Aarhus University, Denmark
association pairs (4, 5). In addition, recent efforts to map the Amyotrophic lateral sclerosis
frameworkBSCL2 known disease gene and phenotype associations.
all Received September 15, 2009; Accepted November 3, 2009; Published November 30, 2009
protein–protein interactions in humans (6, 7), together paraplegia syndrome
Silver spastic
with efforts
ALS2
to curate an extensive map of human metabolism (8) and regulatory Spastic ataxia/paraplegia Copyright: ß 2009 Barrenas et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits
Properties BRIP1the HDN. If each human disorder tends to have a
of
networks offer increasingly detailed maps of theFanconi anemia relationships unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
between different disease genes. Most of the successful studies distinct and unique genetic origin, then the HDN would be dis-
connected into many single nodes corresponding to specific disor- Funding: This work was supported by the Swedish Research Council, The European Commission, The Swedish Foundation for Strategic Research (PH), and the
building on these new approaches have focused, however, on a WCU (World Class University) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology R31-R31-
single disease, using network-based tools to gain a better under- ders or grouped into small clusters of a few closely related disorders. 2008-000-10029-0 (PH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
In contrast, the obtained HDN displays many connections between
Fig. 1. Construction of the diseasome bipartite network. (Center) A small subset of OMIM-based disorder– disease gene associations (18), where circles and rectangles
standing of the relationship between the genes implicated in a
correspond to disorders and disease genes, respectively. A link is placed between a disorder and aindividual disorders and disorder classes (Fig. 2a). Of 1,284
disease gene if mutations in that gene lead to the specific disorder. Competing Interests: The authors have declared that no competing interests exist.
selected circle is proportional to the number of genes participating in the corresponding both and the color corresponds to the disorder class to which the disease
The size of adisorder (9). disorder,
belongs. (Left) The HDN a conceptually different approach, which two disorders are connected there
Here we take projection of the diseasome bipartite graph, in exploring disorders, 867ifhave is a gene that is implicatedother disorders,ofand 516
at least one link to in both. The width * E-mail: fredrik.barrenas@gu.se
a link is proportional to the number of genes and are implicated in both diseases. For example, three genes area giant component, cancer and prostate cancer, genetic
whether human genetic disorders that the corresponding disease disorders form implicated in both breast suggesting that the . These authors contributed equally to this work.
resulting in a link ofrelated to each other at a higher level of cellular andtwo genes are connected if they are involved in the same disorder.shared with other
genes might be weight three between them. (Right) The DGN projection where origins of most diseases, to some extent, are The width of
a link is proportional to the number of diseases with which the two genes are commonly diseases. A full number of genes associated with Fig. 13.
associated. The diseasome bipartite map is provided as SI a disorder, s, has a
organismal organization. Support for the validity of this approach
is provided by examples of genetic disorders that arise from broad distribution (see SI Fig. 6a), indicating that most disorders
relate to a few disease genes, whereas a handful of phenotypes, such Introduction human interactome. A more recent report that evaluated the
a few otherin more than a singlefew phenotypes such as colon Formentary, gene-centered view of the diseasome. Given that the links
mutations disorders, whereas a gene (locus heterogeneity). network properties of disease genes showed that genes with
cancer (linked to k ϭ 50 other disorders) orby mutations (k ϭ 30) atsignify deafnessphenotypicleukemia (s ϭ 37), and colon cancer (s ϭ 34),
example, Zellweger syndrome is caused breast cancer in any of as related (s ϭ 41), association between two genes, they Systems Biology based approaches of studying human genetic intermediate degrees (numbers of neighbors) were more likely to
least 11 hubs that associated with peroxisome biogenesis (10).represent a to dozens of genes (Fig. 2a). The degree (k) distribution of
represent genes, all are connected to a large number of distinct relate measure of their phenotypic relatedness, which could be
diseases have brought in a shift in the paradigm of elucidating harbor germ-line disease mutations [12]. However, interpretation
disorders. The prominence of cancer among the most connected theusedHDN (SI Fig. 6b) in conjunction with protein–proteinlinked to only
Similarly, there are many examples of different mutations in in future studies, indicates that most disorders are inter- disease mechanisms from analyzing the effects of single genes to
disorders arises in part from the many clinically distinct cancercur-actions (6, 7, 19), transcription factor-promoter interactions (20),
same gene (allelic heterogeneity) giving rise to phenotypes of this dataset might not be applicable to complex disease genes
understanding the effect of molecular interaction networks. Such
subtypes tightly connected withdisorders. For example, mutations inand metabolic reactions (8), toM.V., and A.-L.B. designed research; K.-I.G. and M.E.C.
rently classified as different each other through common tumor
Author contributions: D.V., B.C.,
discover novel genetic interactions. since 97% of the disease genes were monogenic. Despite this
TP53 have been linked to 11 clinically distinguishable cancer-In the DGN, research;of 1,777 disease genes data; connected to other M.V., and
repressor genes such as TP53 and PTEN. performed 1,377 K.-I.G. and M.E.C. analyzed are and K.-I.G., M.E.C., D.V.,
networks have been exploited to find novel candidate genes, based reservation, both the latter studies found a functional clustering of
related disorders (11). Given the highly interlinked internal orga-disease genes, and paper.
Although the HDN layout was generated independently of any A.-L.B. wrote the 903 genes belong to a giant component (Fig. 2b). on the assumption that neighbors of a disease-causing gene in a disease genes. Another concern is that the above studies could be
knowledge on disorder classes,itthe resultingpossible to improve theWhereas the number of genesinterest.
nization of the cell (12–17), should be network is naturally The authors declare no conflict of involved in multiple diseases de- network are more likely to cause either the same or a similar confounded by discovery bias, in other words these disease genes
and visibly clustered accordingapproachdisorder classes. Yet, there
single gene–single disorder to major by developing a conceptualcreases article is a (SI Fig. 6d; light gray nodes in Fig. 2b), several
This rapidly PNAS Direct Submission. disease [1–14]. Initial studies investigating the network properties were identified based on previous knowledge. By contrast,
framework to link systematically all genetic disorders (the humandisease genes (e.g., TP53, PAX6) are involved in as many as 10 GO, Gene
are visible differences between different classes of disorders. Abbreviations: DGN, disease gene network; HDN, human disease network; of human disease genes were based on cancers and revealed that
Whereas the large cancer cluster is tightly interconnected due to the(thedisorders, representing major hubs in the network. Pearson correlation coeffi-
‘‘disease phenome’’) with the complete list of disease genes
Genome Wide Association studies (GWAs) do not suffer from
Ontology; OMIM, Online Mendelian Inheritance in Man; PCC, up-regulated genes in cancerous tissues were central in the
many genes associated with multiple global view of (TP53, KRAS,
‘‘disease genome’’), resulting in a types of cancer the ‘‘diseasome,’’ cient.
such bias [15].
ERBB2, NF1, etc.) of all known several diseases with strong pre- **To whom correspondence and DGN. To probe how the topology
the combined set and includes disorder/disease gene associations.Functional Clustering of HDN may be addressed. E-mail: alb@nd.edu or marcvidal@ interactome and highly connected (often referred to as hubs) In this study, we have derived networks of complex diseases and
disposition to cancer, such as Fanconi anemia and ataxia telangi- of thedfci.harvard.edu. GDN deviates from random, we randomly
HDN and [1,2]. A subsequent study based on the human disease network complex disease genes to explore the shared genetic architecture of
Results
ectasia, metabolic disorders do not appear to form a single distinct shuffledarticle contains supporting information online at www.pnas.org/cgi/content/full/
This the associations between disorders and genes, while keep- and disease gene network derived from the Online Mendelian complex diseases studied using GWAs. Further, we have evaluated
cluster but are underrepresented in constructed a bipartite graphing the number of links per each disorder and disease gene in the
Construction of the Diseasome. We the giant component and 0701361104/DC1. Inheritance in Man (OMIM) demonstrated that the products of the topological and functional properties of complex disease genes
overrepresented in disjoint sets of nodes. One set corresponds to allbipartite network unchanged. Interestingly, the average size of the
consisting of two the small connected components (Fig. 2a). To © 2007 by The National Academy of Sciences of the USA disease genes tended (i) to have more interactions with each other in the human interactome by comparing them with essential,
quantify this difference, we measured the locus heterogeneity of giant component of 104 randomized disease networks is 643 Ϯ 16,
each disorder class and the fraction of disorders that are connected significantly larger than 516 (P Ͻ 10Ϫ4; for details of statistical than with non-disease genes, (ii) to be expressed in the same tissues monogenic and non-disease genes. We observed that diseases
www.pnas.org͞cgi͞doi͞10.1073͞pnas.0701361104
to each other in the HDN (see SI Text). We find that cancer and analyses of the results reported hereafter, ͉see SI104 ͉ no. 21 ͉ 8685– 8690
PNAS ͉ May 22, 2007 vol. Text), the actual and (iii) to share Gene Ontology (GO) terms [8]. Contradicting belonging to the same disease class do not always show a tendency
neurological disorders show high locus heterogeneity and also size of the HDN (SI Fig. 6c). Similarly, the average size of the giant earlier reports, this latter study demonstrated that the non-essential to share common disease genes; the complex disease gene net-
represent the most connected disease classes, in contrast with component from randomized gene networks is 1,087 Ϯ 20 genes, human disease genes showed no tendency to encode hubs in the work shows high modularity comparable to that of the human
metabolic, skeletal, and multiple disorders that have low genetic significantly larger than 903 (P Ͻ 10Ϫ4), the actual size of the DGN
heterogeneity and are the least connected (SI Fig. 7). (SI Fig. 6e). These differences suggest important pathophysiological
clustering of disorders and disease genes. Indeed, in the actual PLoS ONE | www.plosone.org 1 November 2009 | Volume 4 | Issue 11 | e8090
Properties of the DGN. In the DGN, two disease genes are connected networks disorders (genes) are more likely linked to disorders
if they are associated with the same disorder, providing a comple- (genes) of the same disorder class. For example, in the HDN there
8686 ͉ www.pnas.org͞cgi͞doi͞10.1073͞pnas.0701361104 Goh et al.](https://image.slidesharecdn.com/eccs-120902-120906011437-phpapp02/85/ECCS12-8-320.jpg)