This document discusses drug resistant tuberculosis (TB) and challenges in diagnosis. It provides an overview of various diagnostic options, including conventional phenotypic methods that take weeks to provide results, as well as newer rapid phenotypic and molecular methods. Line probe assays (LPAs) are highlighted as molecular tests that can identify Mycobacterium tuberculosis and detect genetic mutations associated with resistance to rifampin from cultures or smear positive sputum samples within days, though they are limited in the number of drugs tested. Overall rapid and accurate diagnosis of drug resistant TB is crucial for effective treatment and control.

![Review Article
Department of
Microbiology, Maharaja
Krishna Chandra Gajapati
Medical College and
Hospital, Berhampur,
Odisha, India
Address for correspondence:
Dr. Muktikesh Dash,
E‑mail: mukti_mic@yahoo.
co.in
Drug resistant tuberculosis: A diagnostic
challenge
Dash M
ABSTRACT
Tuberculosis (TB) is responsible for 1.4 million deaths annually. Wide‑spread misuse of anti‑tubercular drugs
over three decades has resulted in emergence of drug resistant TB including multidrug‑resistant TB and
extensively drug‑resistant TB globally. Accurate and rapid diagnosis of drug‑resistant TB is one of the paramount
importance for instituting appropriate clinical management and infection control measures. The present article
provides an overview of the various diagnostic options available for drug resistant TB, by searching PubMed for
recent articles. Rapid phenotypic tests still requires days to weeks to obtain final results, requiring biosafety
and quality control measures. For newly developed molecular methods, infrastructure, training and quality
assurance should be followed. Successful control of drug resistant TB globally will depend upon strengthening
TB control programs, wider access to rapid diagnosis and provision of effective treatment. Therefore, political
and fund provider commitment is essential to curb the spread of drug resistant TB.
KEY WORDS: Diagnosis, drug resistant, rapid, tuberculosis
Received : 10‑10‑2012
Review completed : 03‑12‑2012
Accepted : 07‑06‑2013
Introduction
T he impact of tuberculosis (TB) can be devastating
even today, especially in developing countries suffering
from high burdens of both TB and human immunodeficiency
virus (HIV). In 2010, there were 8.8 million new cases of TB
globally, causing 1.4 million deaths.[1] TB is a major public
health problem in India, which accounts for one‑fifth of
the global TB incident cases. Each year nearly 2 million
people in India develop TB, of which around 0.87 million
are infectious cases.[2] It is estimated that annually around
2,80,000 (23/1,00,000 population) Indians die due to TB.[2]
Drug resistance has enabled it to spread with a vengeance.
The prevalence of multidrug‑resistant tuberculosis (MDR‑TB)
and extensively‑drug resistant TB tuberculosis (XDR‑TB) are
increasing throughout the world both among new TB cases
as well as among previously treated ones.[3] Accurate and
rapid diagnosis of drug‑resistant TB is one of the paramount
importance for instituting appropriate clinical management
and appropriate infection control measures.[4,5] Fortunately, the
past few years have seen an unprecedented level of funding and
activity focused on the development of new tools for diagnosis
of drug resistant TB. This should go a long way in helping us
arrest the spread of the disease.
Sources and method included PubMed search for recent articles
using MeSH terms “TB” and “resistance” and “diagnosis.”
Furthermore, World Health Organization (WHO) reports and
national guidelines were used. The inclusion criteria for selected
articles were based on relevance to the purpose of the review.
Drug Resistant TB
MDR‑TB is a form of TB caused by a strain of Mycobacterium
tuberculosis (MTB) resistant to the most potent first line anti‑TB
drugs, i.e., isoniazid (INH) and rifampicin (RIF). It has been
estimated that India and China account for nearly 50% of the
global burden of MDR‑TB cases.[6] Approximately, 5% of all
pulmonary TB cases in India may be MDR. MDR rates are low
in new, untreated cases. The incidence in such cases ranges
from 1% to 5% (mostly < 3%) in different parts of India.[7,8]
However, during the last decade, there has been an increase in
reported incidences of drug resistance in category II TB cases,
particularly among those treated irregularly or with incorrect
regimens and doses. In such cases, the incidence of MDR‑TB
varies from 11.8% to 47.1%.[9]
XDR‑TB, is defined as TB caused by a strain of MTB that
is resistant to RIF and INH as well as to any member of the
quinolone family and at least one of the second line anti‑TB
Access this article online
Quick Response Code: Website:
www.jpgmonline.com
DOI:
10.4103/0022-3859.118038
PubMed ID:
***
196 Journal of Postgraduate Medicine July 2013 Vol 59 Issue 3](https://image.slidesharecdn.com/drugresistanttuberculosis-140913202334-phpapp02/85/Drug-resistant-tuberculosis-A-diagnostic-challenge-2-320.jpg)
![Dash: Diagnosis of drug resistant tuberculosis
injectable drugs, i.e. kanamycin, capreomycin or amikacin.
XDR‑TB was first described in 2006. Since then, there have
been documented cases in 77 countries world‑wide by the
end of 2011.[10] The global prevalence of XDR‑TB has been
difficult to assess. The prevalence of XDR‑TB has been reported
from India, which varies between low, i.e., 2.4% and as high as
21.1% in HIV infected persons suffering from MDR‑TB.[11,12]
Treatment outcomes are significantly worse for patients with
XDR‑TB, compared with patients with drug‑susceptible TB
or MDR‑TB.[13,14] In the first recognized outbreak of XDR‑TB,
53 patients in KwaZulu‑Natal, South Africa, who were
co‑infected with XDR‑TB and HIV, survived for an average of
16 days, with mortality of 98%.[15] XDR‑TB raises concerns of
a future TB epidemic with restricted treatment options and
jeopardizes the major gains made in TB control.
Totally, drug resistant TB (TDR‑TB) or extremely drug resistant
TB is resistant to all first line and second line anti‑tubercular
drugs. The detection of four Mumbai cases, which were
resistant to all first line and second line drugs.[16] This kind of
rapid progression of drug resistance from MDR, to XDR and
TDR‑TB underlines the need for rapid and accurate diagnosis
of drug resistant TB.
Molecular basis of drug resistance
RIF acts by binding to the beta‑subunit of the ribonucleic
acid (RNA) polymerase (coded for by the rpoB gene),
inhibiting RNA transcription. Subsequent deoxyribonucleic
acid (DNA) sequencing studies have shown that more than
95% of RIF resistant strains have mutations in an 81‑base
pair region (codons 507‑533) of the rpoB gene. INH inhibits
enoyl‑acyl carrier protein (ACP)‑reductase (coded by the inhA
gene), which is involved in mycolic acid biosynthesis. INH is
also a “pro‑drug,” which is converted to its active form by the
catalase‑peroxidase enzyme (coded by katG gene). Resistant
mutants can be due to different regions of several genes,
including binding of activated INH to its inhA target, the
activation of the pro‑drug by katG or by increased expression
of the target inhA. Point mutations in codon 315 of the katG
gene have been found in 50‑90% of high‑level INH resistant
strains while 20‑35% of low‑level INH resistant strains have been
reported mutations in inhA regulatory region and 10‑15% have
mutations in the ahpC‑oxyR intergenic region (often together
with katG mutations in other regions).[17]
Conventional Phenotypic Methods for Diagnosis of Drug
Resistant TB
MTB is an extremely slow growing organism. Using the
standardized drug susceptibility testing (DST) with conventional
methods, 8‑12 weeks are required to identify drug resistant TB
on solid media (i.e. Lowenstein‑Jensen [LJ] medium). In
general, these methods assess inhibition of MTB growth in the
presence of antibiotics to distinguish between susceptible and
resistant strains. As the results usually take weeks, inappropriate
choice of treatment regimen may result in death such as in case
of XDR‑TB (especially in HIV co‑infected patients). In addition,
delayed diagnosis of drug resistance results in inadequate
treatment, which may generate additional drug resistance and
continued transmission in community. The most common
medium for the agar proportion method in resource limited
countries is LJ medium; however, the Clinical and Laboratory
Standards Institute, (CLSI) considers this medium to be
unsuitable for susceptibility testing due to uncertainty about
the potency of drugs following inspissation and also because
components present in the eggs or the medium may negatively
affect some drugs. Both Centers for Disease Control and
Prevention and CLSI recommend that Middle brook 7H10 agar
supplemented with oleic albumin dextrose catalase (OADC)
be used as the standard medium for the agar proportion assay.
Rapid phenotypic methods for diagnosis of drug resistant TB
Rapid automated liquid based culture and susceptibility tests
Automated liquid culture systems such as Bactec radiometric
system (Bactec 460TB; Becton Dickinson, USA), non‑radiometric
systems MB/BacT ALERT (BioMerieux, France), Versa Trek
(Trek Diagnostic System, USA) and mycobacteria growth
indicator tube (MGIT 960; Becton Dickinson, USA) are more
sensitive and shorter turnaround time than solid media cultures.
These are also less labor intensive and therefore, less vulnerable
to manual errors. But, automated systems still requires few
weeks to obtain final results.[18] Also these instruments are costly,
require maintenance and can be extremely difficult for most
public health laboratories in developing countries.
Nitrate reductase assay (Griess method)
The NRA is a liquid or solid medium technique that measures
nitrate reduction by members of the MTB complex to indicate
growth and to indicate resistance to INH and RIF. Since the
NRA method uses nitrate reduction as a sign of growth, results
are detected earlier than by examination of microcolonies on
solid medium. However, the Griess reagent kills the organisms
when added to the tubes, so multiple tubes must be inoculated
if further testing is necessary. In addition, not all members of the
MTB complex reduce nitrate, so the presence of nitrate‑negative
acid fast bacilli (AFB) may require further testing.
Thin layer agar cultures and TK medium
Thin layers of middle brook 7H11 solid agar medium are used
to detect microcolonies by conventional microscopy. It can
be adapted for the rapid detection of drug resistance directly
from sputum samples, but requires average turnaround time
of 11 days.[19]
Newly developed test such as TK medium (Salubris Inc., USA)
is a colorimetric system that indicates growth of mycobacteria
by changing the color of the growth medium. Metabolic activity
of growing mycobacteria changes the color of the culture
medium and this allows for an early positive identification before
bacterial colonies appear. Unfortunately, there is insufficient
published evidence on the field performance of these tests in
developing countries.[20]
Microscopic observation drug susceptibility assay
The MODS assay is based on characteristic cord formation
of MTB that can be visualized microscopically (“strings
and tangles” appearance) in liquid medium with or without
antimicrobial drugs (for DST).[21] The test sensitivity is better
Journal of Postgraduate Medicine July 2013 Vol 59 Issue 3 197 ](https://image.slidesharecdn.com/drugresistanttuberculosis-140913202334-phpapp02/85/Drug-resistant-tuberculosis-A-diagnostic-challenge-3-320.jpg)
![Dash: Diagnosis of drug resistant tuberculosis
than traditional methods using LJ media with a turnaround time
of 7 days for culture and drug susceptibility testing (C‑DST) for
INH and RIF. It is cheap, simple and fairly accurate.[22] Biosafety
level‑3 facilities are required if the plates are opened for further
testing, such as for the confirmation of the identification of
TB. Laboratories using MODS assay require a functioning
biosafety cabinet, a safety centrifuge, an incubator, an inverted
light microscope for observation of mycobacterial growth and
supplemented liquid media (Middle brook 7H9 broth with
OADC and PANTA).
Phage based assay
Phage amplification‑based test (FAST Plaque‑Response,
Biotech Laboratories Ltd. UK) has been developed for direct
use on sputum specimens. Drug resistance is diagnosed when
MTB is detected in samples that contain the drug (i.e., RIF).
When these assays performed on MTB culture isolates, they
have shown high sensitivity and variable specificity, but the
evidence is lacking about the accuracy when they are directly
applied to sputum specimens.[23] It also requires high standards
of biosafety and quality control.
Luciferase reporter phages are genetically‑modified phages
containing the flux gene encoding firefly luciferase. This
catalyzes a reaction producing light in the presence of the
luciferin substrate and adenosine triphosphate; light is only
produced in the presence of viable mycobacteria. Detection
of light released from viable mycobacteria can be achieved
by a luminometer or photographic film within 2‑4 days from
culture.[24] The luminometer readout is more sensitive and
enables quantification of results while the use of Polaroid
photographic film offers a lower‑tech approach with lower
sensitivity.
Rapid molecular methods for diagnosis of drug resistant TB
Since the publication of genome details of MTB H37Rv strain
in 1998, these have been utilized in development of nucleic
acid amplification (NAA) tests for diagnosis of drug resistant
TB. A number of NAA tests are now available, manual and
automated, commercial and in the house, with varying
performance characteristics. Real‑time polymerase chain
reaction (RT‑PCR) and line probe assays (LPAs) have been
commercialized and widely used in clinical laboratories merit
special mention, detailed below.
RT‑PCR
Molecular tools are based on identification of specific mutations
responsible for drug resistance, which are detected by the process
of NAA in conjunction with electrophoresis, sequencing or
hybridization. Direct sequencing techniques such as RT‑PCR
that uses wild‑type primer sequences to amplify genes and
enable the use of specific probes to identify mutations. Recently
introduced semi‑quantitative nested RT‑PCR, which integrates
and automates sample processing and simultaneously detects
MTB and RIF resistance within the single‑use disposable
cartridges. A study examined 1,730 patients with suspected
drug‑sensitive or MDR pulmonary TB across Peru, Azerbaijan,
South Africa and India. There was sensitive detection of MTB
and RIF resistance directly from untreated sputum in < 2 h with
minimal hands‑on time.[25] The WHO has recently supported
the use of this system as an initial diagnostic test in respiratory
specimens of patients with high clinical suspicion of having TB
or who could be MDR.[26] These tests are very expensive require
adequate maintenance and calibration of the equipment.
Further, the specificity for RIF resistance in populations where
MDR‑TB is rare requires confirmation by repeat testing and
thus increases cost.
LPAs
LPAs are a family of novel DNA strip tests that use both PCR and
reverse hybridization methods. In these assays, a specific target
sequence is amplified and applied on nitrocellulose membranes.
Specific DNA probes on the membrane hybridize with the
amplified sequence applied on it. Color conjugates make the
amplified target sequences appear as colored bands. These tests
have been designed to identify MTB and simultaneously detect
genetic mutations related to drug resistance both from clinical
samples as well as culture isolates. These tests able to identify
MTB complex and simultaneously detect genetic mutations in
the rpoB gene region related to rifampin resistance. The LPA
strip consists of 10 oligonucleotide probes: One is specific for
the MTB complex, five are partially overlapping wild‑type probes
that span the region at positions 509‑534 of the rpoB gene and
four probes are specific for amplicons carrying the most common
rpoB mutations (D516V, H526Y, H526D and S531L). According
to a recent review, the sensitivities of the LPA are above 90% for
clinical isolates with 99‑100% specificity [Table 1]. It identifies
MTB complex and simultaneously detects mutations in the rpoB
gene as well as mutations in the katG gene for high‑level INH
resistance. These tests can also detect mutations in the inhA gene
for low‑level INH resistance and mutations in the gyrA, rrs and
embB genes for 2nd line anti‑tubercular drugs fluoroquinolones,
aminoglycosides and ethambutol respectively [Table 2].[32] The
newly developed assay may represent a reliable tool for detection
of fluoroquinolones, amikacin, capreomycin and ethambutol
resistance. LPA strip can use culture isolates and smear positive
sputum as specimen and provide results in 1‑2 days. A recent
laboratory evaluation study from South Africa estimated the
accuracy of the GenoType MTBDRplus assay performed directly
on AFB smear‑positive sputum specimens.[33] It showed high
sensitivity, specificity, positive and negative predictive values
for detection of RIF and INH resistance [Table 1]. However,
a meta‑analysis on this assay found that sensitivity estimates
for INH resistance were comparatively modest.[33,34] In general,
LPAs are expensive and require dedicated equipments, reagents
and facilities. Molecular drug resistance testing has technical
limitations, i.e., LPAs can detect only well characterized
drug resistance alleles, not every drug resistance allele can be
discriminated via current tests and silent mutations (which do
not confer drug resistance) can be detected by probes leading
to misclassification of drug resistance. In addition, molecular
tests cannot determine the proportion of drug resistant bacteria
within a mixed population of cells (i.e., wild type and drug
resistant). Cross contamination with amplicons generated from
previous tests can be problematic especially when the tests have
been employed in laboratories without appropriate staff training
and quality control.
198 Journal of Postgraduate Medicine July 2013 Vol 59 Issue 3](https://image.slidesharecdn.com/drugresistanttuberculosis-140913202334-phpapp02/85/Drug-resistant-tuberculosis-A-diagnostic-challenge-4-320.jpg)
![Dash: Diagnosis of drug resistant tuberculosis
Table 1: Performance of methodologies used to diagnose drug resistant tuberculosis
Drug resistance Methodology and products Sensitivity % (95% CI) Specificity % (95% CI) Reference number
Phenotypic methods
Automated liquid
MB/BacT
for rifampicin
based CST systems
ALERT
resistance
Versa TREK 100 100
MGIT 960 99.8 99.2
NRA Griess method 99 100 [28]
MODS 98 (94.5‑99.3) 99.4 (95.7‑99.9) [29]
Phage based assay FASTPlaque‑response
(commercial)
FASTPlaque‑Response
(in‑house)
Luciferase reporter
phage
Phenotypic methods
for isoniazid
resistance
Automated liquid
based CST systems
MB/BacT ALERT 100 95 [27]
Versa TREK 95.5 99
MGIT 960 97.1 100
NRA Griess method 94 100 [28]
MODS 97.7 (94.4‑99.1) 95.8 (88.1‑98.6) [29]
Molecular methods
for rifampicin
resistance
RT‑PCR GeneXpert® MTB/RIF 98 (97‑99) 99 (98‑99) [31]
Line probe assay INNO‑LiPA Rif. TB 93 (89‑96) 99 (99‑100)
MTBDRplus 97 (92‑99) 98 (95‑99)
Molecular methods
for isoniazid
resistance
Line probe assay MTBDRplus 77 (69‑83) 99 (97‑100)
Molecular methods
for second‑line drug
resistance
Line probe assay MTBDRsl
(fluoroquinolones)W
Amikacin 90 (81‑96) 100 (98‑100)
Kanamycin 83 (59‑96) 100 (96‑100)
Capreomycin 87 (77‑94) 99 (96‑100)
Ethambutol 60 (52‑68) 98 (94‑100)
CST – Culture and susceptibility testing; NRA – Nitrate reductase assay; MODS – Microscopic observation drug susceptibility; RT‑PCR – Real time
polymerase chain reaction; CI – Confidence interval; MTB – Mycobacterium tuberculosis; TB – Tuberculosis; RIF – Rifampicin; MGIT – Mycobacteria
growth indicator tube
Diagnostic testing algorithm for drug resistant TB
Laboratory policies and testing of patients suspected of
having drug‑resistant TB depend on the local epidemiology,
local treatment policies, the existing laboratory capacity,
specimen referral and transport mechanisms and human and
financial resources. Mycobacterial culture (solid or liquid)
and identification of MTB provide a definitive diagnosis of
TB. Culture also provides necessary isolates for conventional
DST. Thus LPAs, RT‑PCRs, MODS and NRA can be used in
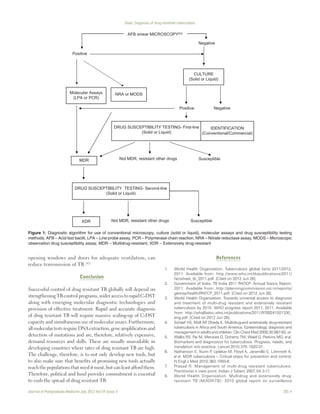
conjunction with culture (solid and liquid) and DST [Figure 1].[35]
Revised national tuberculosis control program and diagnosis
of drug resistant TB
The RNTCP plans to strengthen laboratory capacity for MTB
C‑DST and LPA across India. To date, 35 RNTCP accredited
labs including 14 LPA and 4 liquid culture labs in public and
private sectors are serving patients while another 30 labs are
under the process of up‑gradation and accreditation under
RNTCP, most of them include LPA and liquid culture for first
and second line drugs.[36] In a policy statement released in June
2008, the WHO endorsed the use of LPA for rapid screening
of patients at risk of MDR‑TB and recommended the use
99 98 [27]
95.5 (92.2‑97.4) 95 (91.3‑97.2) [30]
98.5 (96.1‑99.4) 97.9 (94.8‑99.2)
99.3 (49.1‑100) 98.6 (92.5‑99.8)
85 (78‑91) 100 (97‑100)
of LPAs only on culture isolates and smear‑positive sputum
specimens. It is not recommended as a complete replacement
for conventional C‑DST.[37] As of January 2012, diagnosis of
XDR‑TB can only be confirmed at three laboratories in India,
which are quality assured for second line anti‑TB DST of
flouroquinolones and injectable drugs. These are the National
Reference Laboratories (NRL) of tuberculosis research centre/
national institute for research in tuberculosis (TRC/NIRT)
Chennai, National Tuberculosis Institute (NTI) Bangalore
and LRS Institute, New Delhi. Routine fluoroquinolone and
injectable DST (i.e., XDR‑TB diagnosis) on all MDR‑TB
patients at the beginning of treatment has been recommended
by the RNTCP National Laboratory Committee in 2011, but
the capacity to conduct that testing is not yet present in most
C‑DST laboratories used by RNTCP. Capacity building for
second line DST is being undertaken through these NRLs.[26]
Prevention and Control of Drug Resistant TB
The principles of TB control are important for the prevention
of drug resistant TB; these include prompt case detection,
provision of curative treatment and prevention of transmission.
Journal of Postgraduate Medicine July 2013 Vol 59 Issue 3 199 ](https://image.slidesharecdn.com/drugresistanttuberculosis-140913202334-phpapp02/85/Drug-resistant-tuberculosis-A-diagnostic-challenge-5-320.jpg)
![Dash: Diagnosis of drug resistant tuberculosis
Table 2: Summary of technologies used to diagnose drug resistant tuberculosis
Technologies Summary of technologies Estimated costs
Description Product Training Infrastructure Equipment Consumable
New solid
culture
methods
Measures nitrate reduction
to indicate RIF and INH
resistance
Non‑commercial (nitrate
reductase assay)
Simultaneously detects TB
and indicate RIF and INH
resistance
Non‑commercial (thin layer
agar culture)
Liquid culture Broth based manual and
automated culture system;
can be configured for DST
Commercial
BacT/ALERT
Versa TREK
MGIT 960
MODS Manual liquid culture
technique uses inverted
microscope to detect TB
Non‑commercial Extensive BSL 2 to
RT‑PCR Allows automated
sample processing, DNA
amplification detection of
TB and screening for RIF
resistance
Commercial (GeneXpert®
MTB/RIF)
GeneXpert detect mutations
in 81‑base pair core region
of rpoB and in codon 315
of katG genes respectively
Molecular line
probe assay
Strip test simultaneously
detects TB genetic
mutations for RIF/INH
resistance
Commercial (INNO‑LiPA,
MTBDR, MTBDRplus,
MTBDRsl)
INNO‑LiPA targets the
16S‑23S DNA spacer
region for mycobacteria
and mutations in the rpoB
gene region related to RIF
resistance
MTBDR detect the
23S rRNA genes for
mycobacteria and
mutations in rpoB as well
as katG genes for INH
resistance
MTBDRplus target in
addition inhA for low level
INH resistance
MTBDRsl detects in
addition gyrA, rrs and
embB for fluoroquinolones,
aminoglycosides and
ethambutol respectively
RIF – Rifampicin; INH – Isoniazid; MODS – Microscopic observation drug susceptibility; RT‑PCR – Real time polymerase chain reaction;
TB – Tuberculosis; DST – Drug susceptibility testing. Basic laboratory (no specialized biosafety equipment). BSL 2 – Biosafety level 2 (specialized
biosafety equipment required, such as biosafety cabinet); BSL 3 – Biosafety level 3 (biosafety cabinet and other safety equipment required.
Controlled ventilation system that maintains a directional airflow into the laboratory also required); MGIT – Mycobacteria growth indicator tube;
MTB – Mycobacterium tuberculosis; DNA – Deoxyribonucleic acid; rRNA – Ribosomal ribonucleic acid
The WHO’s “Stop TB”[38] directly observed treatment short
course (DOTS) strategy includes supervision and support
of treatment, although there is little evidence that directly
observed treatment alone improves cure rates.[39] Ineffective
drug treatment is a strong risk factor for acquired drug resistance
and proper administration of anti‑tubercular drugs is critical
to reduce this risk. An enhanced DOTS program, DOTS‑plus
has been developed for managing MDR‑TB in resource limited
settings.[40] This program recommends additional facilities for
C‑DST for detection of drug resistant TB and provision of
appropriate second line anti‑tubercular drugs.
Moderate BSL 2 to
BSL 3
Low Medium
Extensive BSL 2 to
BSL 3
Low Medium
Extensive
(3 weeks)
BSL 3 High High
BSL 3
Medium Medium
Minimal Basic
laboratory
High High
Moderate
(3 days)
BSL 2 to
BSL 3
High High
As most of the resistance arises from either inadequate or
inappropriate treatment of active disease, prevention of
active disease indirectly prevents drug resistance. Contact
tracing including family members and health‑care workers,
those who are at risk of acquiring drug resistant TB[41] is one
of the fundamental measures to detect active disease, must
be aligned to all health services. Prevention of transmission
in healthcare settings is difficult in places where resources
are limited with no isolation facilities; one approach is to
manage cases with similar resistance profiles in segregated
groups. However, simple, low‑cost interventions, such as
200 Journal of Postgraduate Medicine July 2013 Vol 59 Issue 3](https://image.slidesharecdn.com/drugresistanttuberculosis-140913202334-phpapp02/85/Drug-resistant-tuberculosis-A-diagnostic-challenge-6-320.jpg)